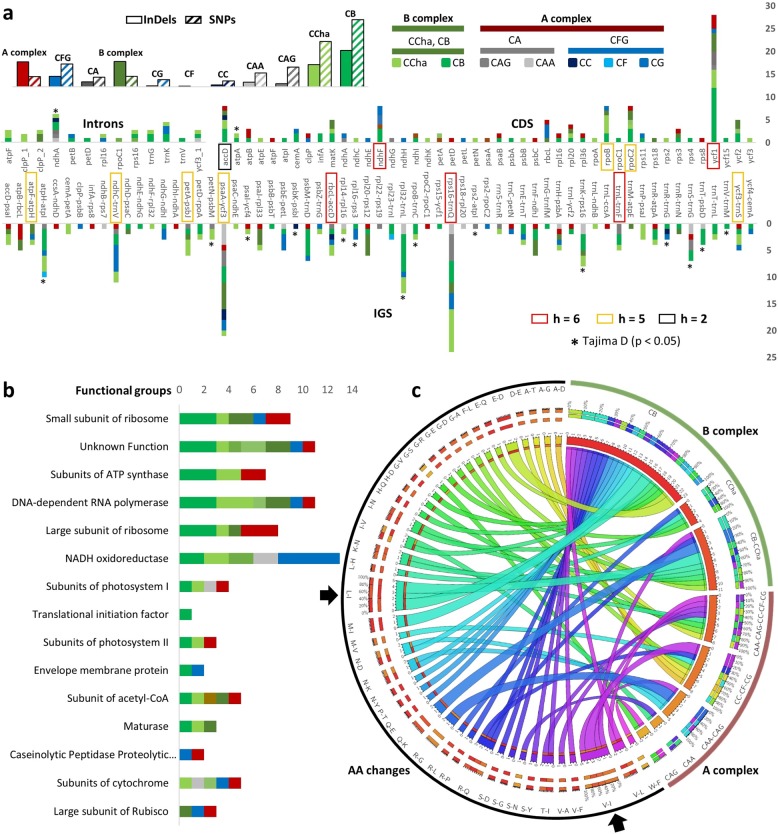
Fig. 3. SNP-based genetic variation in each cultivated Capsicum species/variety.
a Collective stacked column plots for the number of SNP-based polymorphic sites and InDel-based polymorphic sites recorded for each species/variety/group are shown; unique and common SNP counts for every intron and CDS (columns up) and IGS (columns down) per species/variety/group, unneutral loci were marked by an asterisk (*) while neutral highly polymorphic loci were boxed in a colored box (red: haplotypes number h = 6, orange: h = 5, and black: h = 2). b Common and unique SNPs recorded for each functional group per species/variety/group, where the NADH accumulated the highest number of SNPs. c Circos plot shows the number of unique and common amino-acid changes per species/variety/group; where L > I and V > I (indicated by black arrows) are the two most frequent common amino-acid changes for the B complex and A complex, respectively