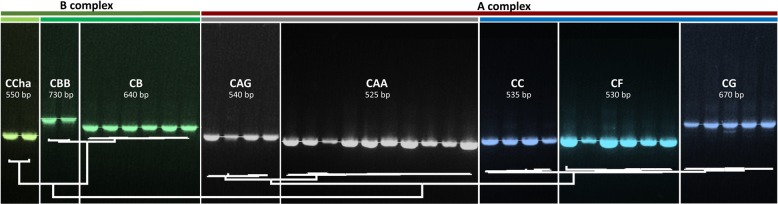
Fig. 4. Colored and sized agarose gel electrophoresis shows rpl23-trnI loci amplified for representative non-gapped haplotypes of the studied Capsicum species/varieties.
The amplified product was polymorphic and capable of discriminating the two complexes between 750–500 bp. Within the A complex (indicated by the red upper line), the product size showed a very narrow difference (~5–10 bp), except for CG, which shows a longer product of 670 bp; the CA species is indicated by a gray upper line, and the CFG clade is indicated by a blue upper line. Within the B complex (defined by dark-green underline), CCha (550 bp) is clearly smaller than CB (730–640 bp; defined with green underline) in product size. A clear difference was observed within the CB species: the highest in product size were CBB by label, while the low product sizes were of different varieties including some labeled as CBB. The product size differences reflect the same highly supported phylogenetic signal estimated by FastTree (bootstrap support >0.8) based on the variable sites of the whole plastome of the representative non-gapped haplotypes