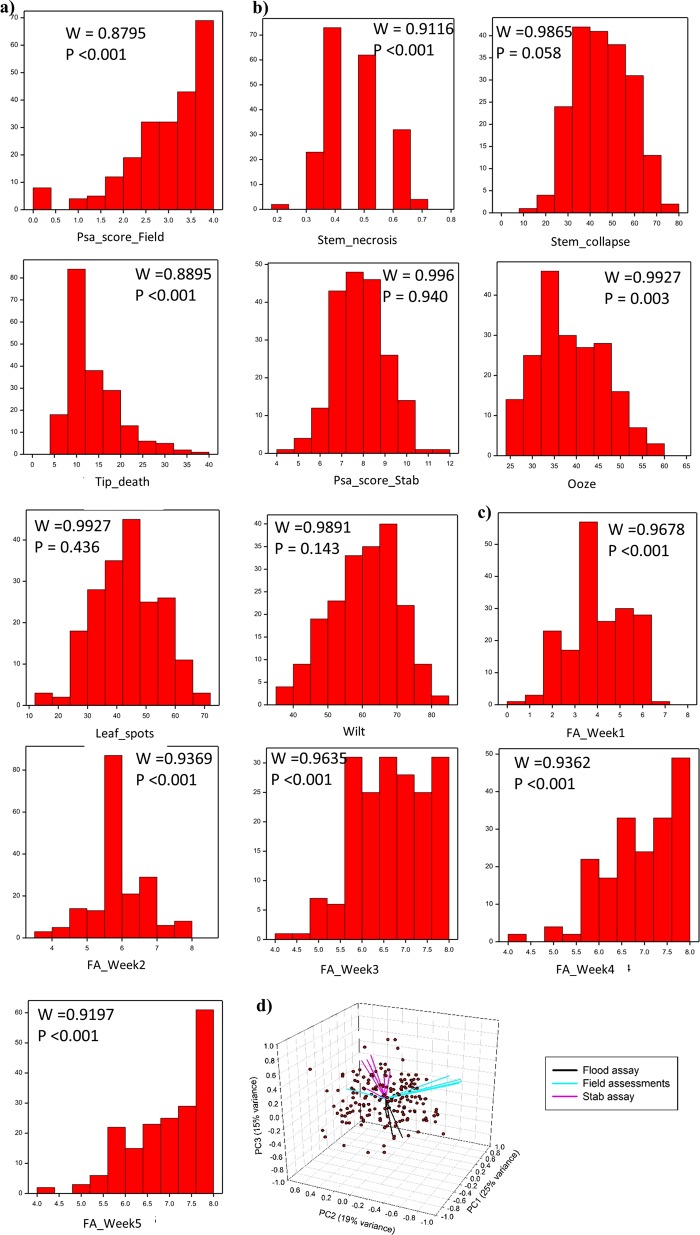
Fig. 2. Distribution of phenotypes in the ‘Hort16A’ × P1 mapping population of 236 genotypes.
a Least squares mean (LSM) of Psa_score_Field after 15 months in the field. The x-axis displays the progression of susceptibility from left to right, while the y-axis represents frequency in the population for the distribution of the trait on the x –axis. b LSM of phenotypes from the stab assay, including Stem_necrosis, Stem_collapse, Tip_death, Psa_score_Stab, Ooze, Leaf_spot, and Wilt. c Means of the health score from Flood bioassays (FA_Week 1 to FA_Week5). The WSTATISTIC is from the Shapiro-Wilks test for the null hypothesis that the distribution is normal. Phenotypic scores with P < 0.001 are rejected for the hypothesis that these distributions are normal. d Principal components analysis on the correlation matrix of the field assessment, flood assay and stab assay measures. Genotypes are shown as points and measurements are shown as vectors (lines pointing from the origin) defined by their correlation with the three principal components