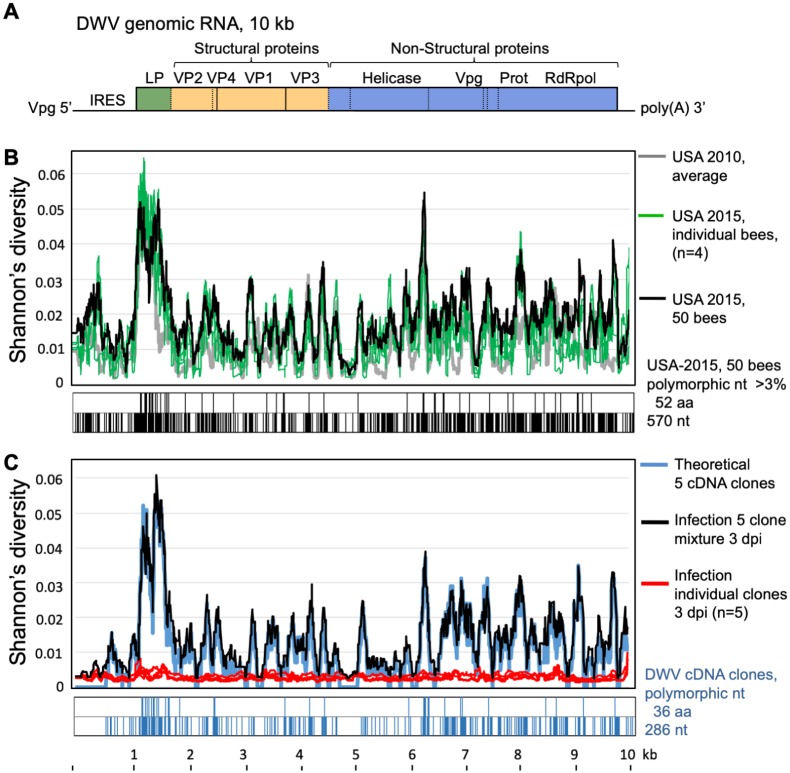
Fig 1. Polymorphisms in natural and clone-derived DWV.
(A) Reference map of DWV genomic RNA. (B,C) Shannon's diversity profiles for DWV (averaged for 100 nt) for (B) 2010 bees (gray line), 2015 individual honey bees (green lines), and a colony sample (black line); (C) individual bees infected with cDNA clone-derived DWV isolates—red lines, single infections; black line, mixed infection of five clones; blue line, theoretical profile for five clone mixture. Distributions of polymorphic nucleotides showing an alternate allele exceeding 3% in frequency and the resulting amino acid changes are shown below the diversity graphs. NGS libraries’ IDs (S1 Table) for the groups are shown in brackets: “USA 2010, average” (B2_PBS, B5_PBS, G2_PBS, G5_PBS, B2_Hem, B5_Hem, G2_Hem, G5_Hem), “USA 2015, individual bees” (V52_Pupa15, V56_Pupa15, V57_Pupa15, V62_Pupa15), “USA 2015, 50 bees” (V99_VIROCT15), “Infection 5 clone mixture 3 dpi” (V112_ABCDE_3), and “Infection individual clones 3 dpi” (V22_304TR, V25_306TR, V16_422TR, V19_702TR, V21_703TR). dpi, days postinoculation; DWV, deformed wing virus; ID, identifier; IRES, internal ribosome entry site; LP, leader protein; NGS, next-generation sequencing; Prot, viral protease; RdRpol, RNA-dependent RNA polymerase; VP, structural viral protein; Vpg, genome-linked protein.