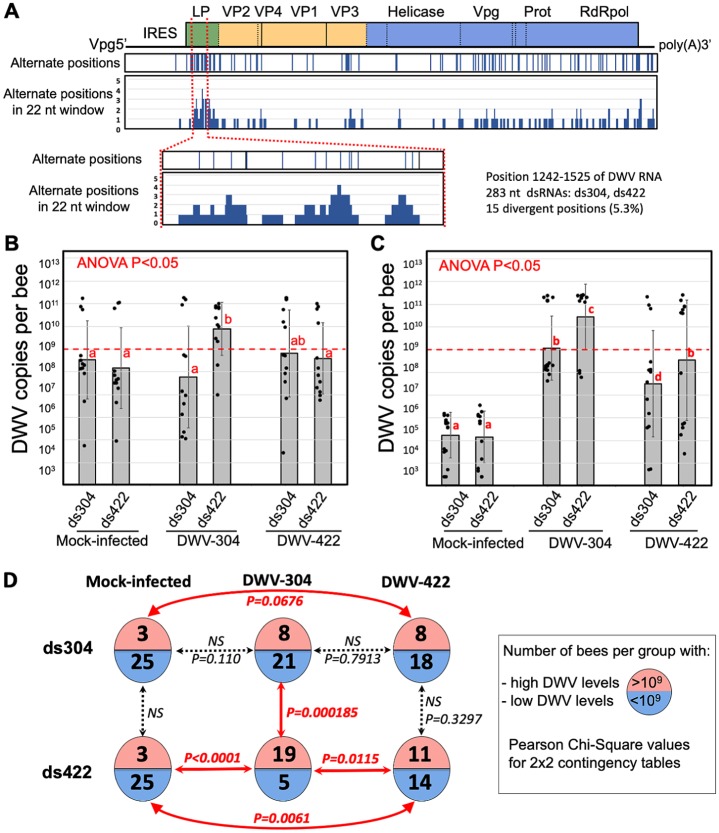
Fig 7. Impact of specificity of RNAi targeting on outcome of DWV infections.
(A) Design of the 283-nt dsRNA specific to the DWV isolates 304 and 422 (ds304 and ds422) corresponding to the regions 1,242–1,524 of DWV genome (GenBank accession MG831200). (B, C) DWV loads in the individual adult honey bees 7 days postfeeding with 108 clone-derived virus particles and 1 μg of the DWV-304- or DWV-422-specific dsRNA. Columns show average copy number per treatment groups ± SD; black dots indicate DWV copy numbers in individual honey bees. The groups with significantly different (P < 0.01, ANOVA) DWV copy numbers are indicated above the bars with different letters. The red lines indicate the threshold level of 109 DWV genome copies per honey bee, which was used to categorize DWV levels as low or high. Numerical values underlying the summary graphs are provided in S1 Data. (D) Analysis of the DWV infection outcomes in two oral inoculation experiments with the strain-specific dsRNAs. Circles indicate experimental groups; numbers of the bees in each group with high (>109 copies) or low DWV (below 109 DWV copies) levels of DWV are shown on the top or bottom of the circles. A 2 × 2 contingency table analysis was used to test significance of the differences in proportions of the bees with low or high DWV levels. dsRNA, double-stranded RNA; DWV, deformed wing virus; IRES, internal ribosome entry site; LP, leader protein; NS, not significant; Prot, viral protease; RdRpol, RNA-dependent RNA polymerase; RNAi, RNA interference; VP, structural viral protein; Vpg, genome-linked protein.