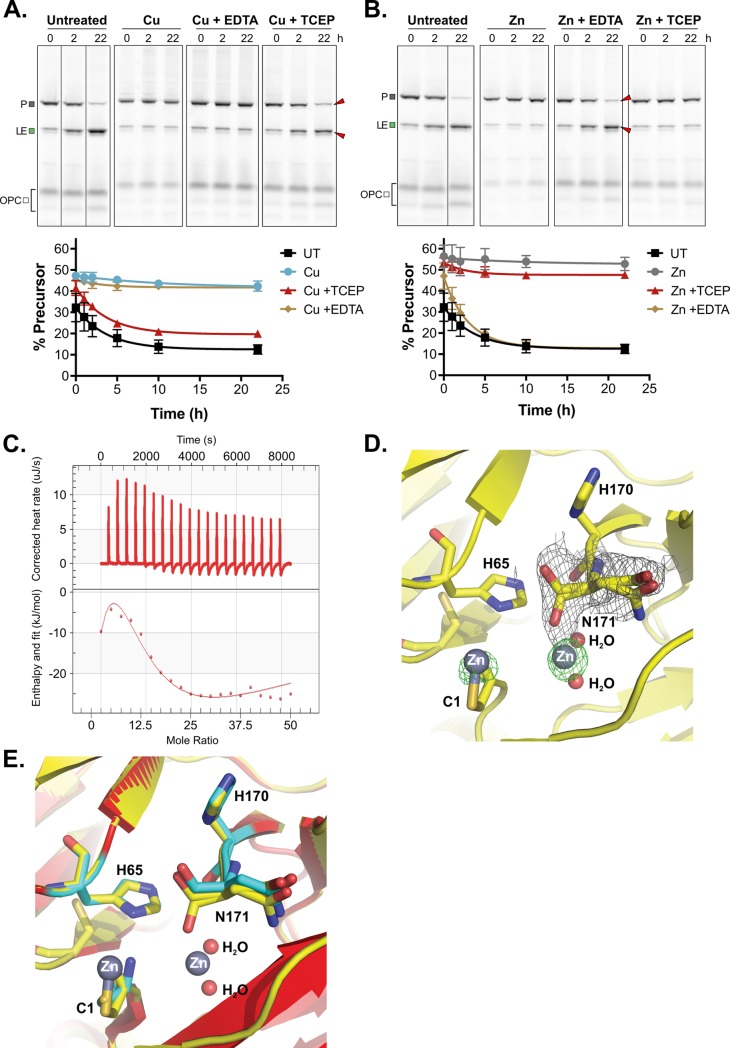
Fig 5. MIG Prp8 A-1V is differentially inhibited by copper and zinc.
(A) Copper inhibition is alleviated by reducing agent only. MIG Prp8 A-1V splicing was completely inhibited by 1 mM copper treatment (Cu) over 22 h, given minimal loss in P or increase in LE occurred compared with the UT control. The inhibition was unaffected by treatment with metal chelator EDTA at 10 mM (Cu + EDTA). Upon adding copper for 2 h and then reducing agent TCEP at 40 mM (Cu + TCEP), splicing was restored and P converted into LE over time. Red arrows indicate splicing rescue. The splice products were quantitated, and the percent precursor is plotted as a proxy for splicing inhibition. Representative gels are shown. Data are representative of 3 biological replicates and mean standard deviations are shown. Data available in S1 Data. Lines through gels indicate where intervening lanes were cropped out of the image. (B) Zinc treatment is relieved by EDTA only. MIG Prp8 A-1V splicing was strongly inhibited by 1 mM zinc treatment (Zn) over 22 h compared with UT lysates. The zinc-based inhibition was relieved when treated with 10 mM EDTA (Zn + EDTA) after 2 h of zinc treatment, and splicing was observed at a rate comparable with the untreated samples. Red arrows indicate splicing rescue. When adding zinc for 2 h and then reducing agent TCEP at 40 mM (Zn + TCEP), splicing was unaffected. Representative gels are shown. Data are representative of 3 biological replicates and mean standard deviations are shown. Data available in S1 Data. Lines through gels indicate where intervening lanes were cropped out of the image. (C) Zinc binds to the Cne Prp8 intein tightly. Using ITC, 16 μM purified Cne Prp8 intein was titrated with 0.05 mM ZnSO4 over 20 injections at 37°C and pH 7.0 on a Nano ITC. The binding isotherm (bottom) shows integrated heat per mole of ZnSO4 as a function of the molar ratio of ZnSO4 to the Cne Prp8 intein and a Kd of 1 ± 0.82 nM was calculated. The NanoAnalyze ITC software automatically discarded outlier data points. Experiment was performed in triplicate. (D) Two binding sites in the Cne Prp8 intein-Zn2+ crystal structure. A close-up view of the crystal structure of the Cne Prp8 intein soaked with zinc shows 2 densities, one surrounding the terminal asparagine (N171) at the C terminus and one around the catalytic cysteine (C1), at the N terminus. Electron density maps are shown for the bound Zn2+ with an omit Fo-Fc difference map (green mesh) contoured at 5δ level and for the alternative conformation of N171 with a 2Fo-Fc map (gray mesh) contoured at 1δ level. Atomic colors are as follows: oxygen, red; carbon, yellow; nitrogen, blue; Zn2+, gray. Zn2+ and water molecules are shown as spheres, and the Cne Prp8 intein residues at the binding site are represented as sticks. (E) Minor conformational changes in the zinc-bound Cne Prp8 intein. A structural superimposition is shown of the native Prp8 intein (red) and the Prp8-Zn2+ complex (yellow) at the Zn2+ binding sites. Atomic colors are as in panel D, except that the carbon atoms for residues C1, H65, H170, and N171 of the native structure are in cyan. Cne, C. neoformans; EDTA, ethylenediaminetetraacetic acid; ITC, isothermal titration calorimetry; LE, ligated exteins; MIG, MBP-Intein-GFP; P, precursor; Prp8, pre-mRNA processing factor 8; TCEP, tris-(2-carboxyethyl)phosphine; UT, untreated.