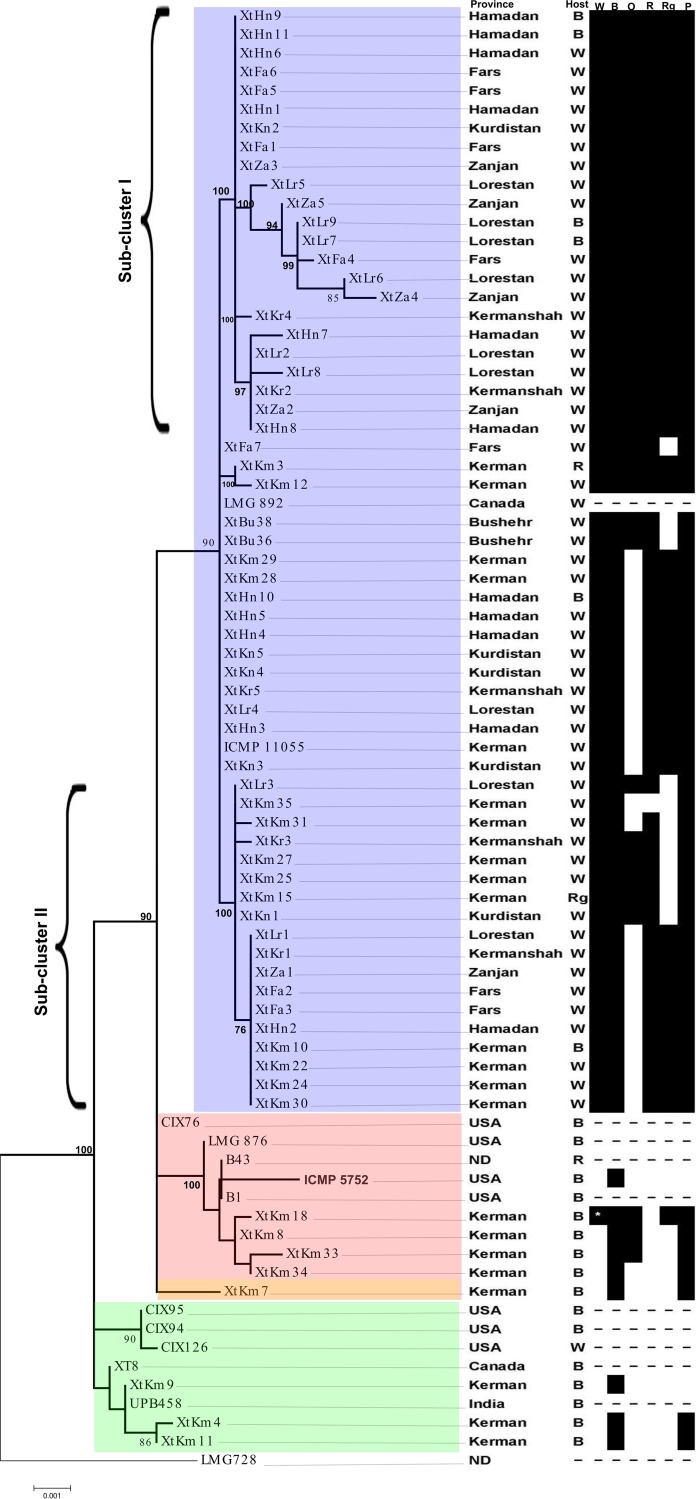
FIG 1.
Phylogeny of Xanthomonas translucens strains isolated in Iran based on the concatenated sequences of four housekeeping genes (dnaK, fyuA, gyrB, and rpoD). Xanthomonas translucens pv. poae (LMG 728T) was used as an out-group to root the tree. The bar presents numbers of substitutions per site. Among the 65 Iranian strains, 57 strains were identified as X. translucens pv. undulosa, while 8 strains were identified as X. translucens pv. translucens. The blue background indicates X. translucens pv. undulosa strains, while red, yellow, and green backgrounds indicate different subclusters of X. translucens pv. translucens. The black-and-white table at the right shows hosts of isolation (Host) and host ranges of the strains on six gramineous species, including wheat (W), barley (B), oat (O), rye (R), ryegrass (Rg), and Harding’s grass (P [Phalaris]), where black squares indicate pathogenicity and white squares indicate nonpathogenicity. A dash mark in the host range table means that pathogenicity has not been determined. The atypical strain XtKm18 (indicated with an asterisk inside a black square), which was isolated from barley and identified as X. translucens pv. translucens, induced water-soaking symptoms on wheat plants.