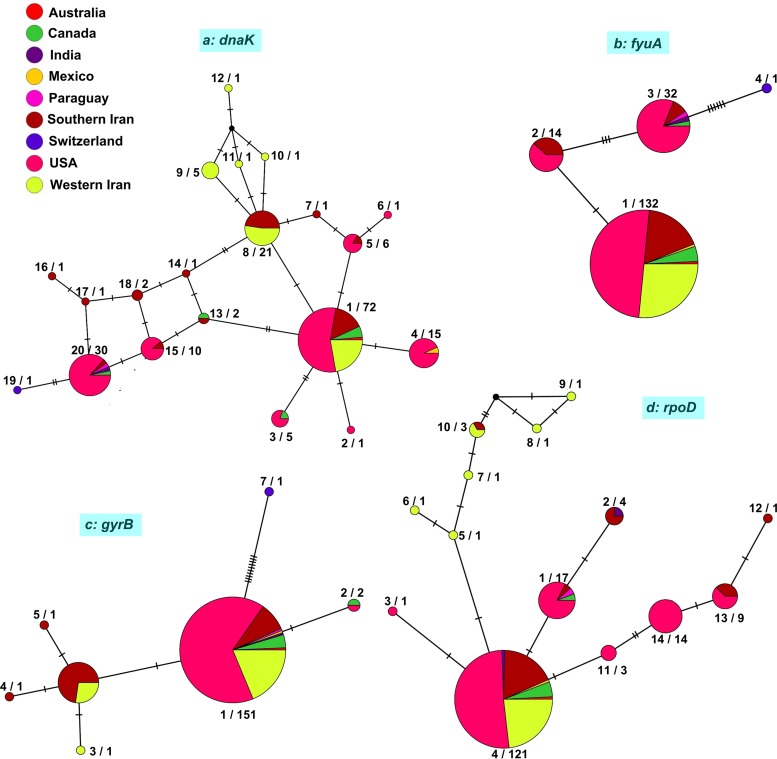
FIG 3.
TCS haplotype network generated using the POPART program from the partial sequences of four housekeeping genes (dnaK, fyuA, gyrB, and rpoD) in 178 Xanthomonas translucens strains. The size of a circle indicates the relative frequency of sequences belonging to a particular sequence type. Hatch marks along the branches indicate the numbers of mutations. Each color indicates a different geographic area. To provide precise insight into the diversity of the pathogen, strains isolated in southern Iran and western Iran were considered distinct groups. The number to the left of a slash represents the ST number (Table S1), while the number to the right of a slash indicates the number of strains in a given ST. Strains isolated in Iran generated several unique haplotypes, indicating a higher genetic diversity of the pathogen in Iran.