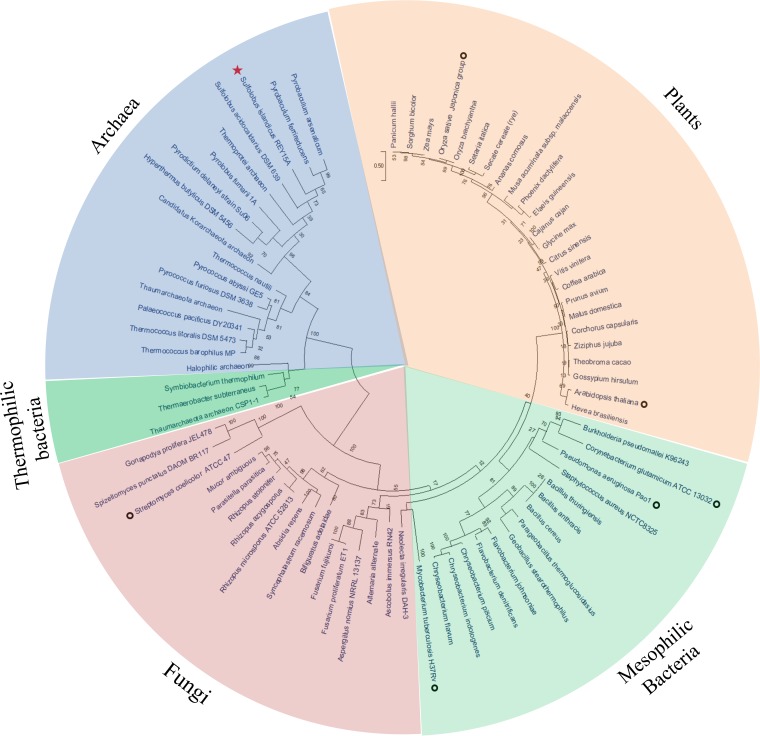
FIG 9.
Phylogenetic tree of LOG proteins from the three domains of life. The unrooted maximum-likelihood method (40) was used to generate a phylogenetic tree with the MEGA X server (39). The circular model tree comprised 76 representative LOG homologue sequences from eukaryotes, bacteria, and archaea (including four uncultured groups, Thermoprotei archaeon, Thaumarchaeota archaeon, “Candidatus Korarchaeota” archaeon, and halophilic archaeon), and all amino acid sequences from Fig. 1 are included. Bootstrap values are indicated at each junction (node) as percentages. The red star indicates S. islandicus REY15A strain used in the present study, and open black circles indicate strains from eukaryotes (8, 10, 11) and bacteria (21, 26, 28, 30) for which phosphoribohydrolase activities have been reported.