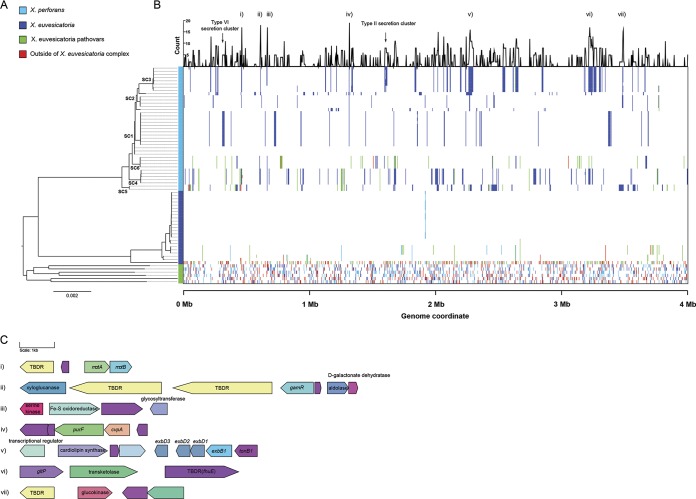
FIG 4.
Landscape of homologous recombination within the X. euvesicatoria species complex. (A) Phylogeny of 67 X. perforans, X. euvesicatoria, and closely related X. euvesicatoria pathovars based on a core genome alignment of 3.98 Mb. The branches corresponding to the sequence clusters inferred within X. perforans (Fig. 1) are labeled at the nodes, and lineages predicted by the FastGear software are color coded at the right of the tree. (B) Patterns of recent recombination across the core genome of the X. euvesicatoria species complex. The colors indicate the donor lineages of the recent recombination events, and the line graph above shows the frequency of recombination within X. perforans at a particular site in the alignment. Recombination events originating from outside the sampled Xanthomonas population are shown in red. (C) Gene contents of core genome loci with elevated recombination frequencies (numbered i to vii) within X. perforans. Genes are color coded according to their annotation, with hypothetical proteins shown in purple. Gene names/functional annotations are shown where available. TBDR, putative TonB-dependent receptor.