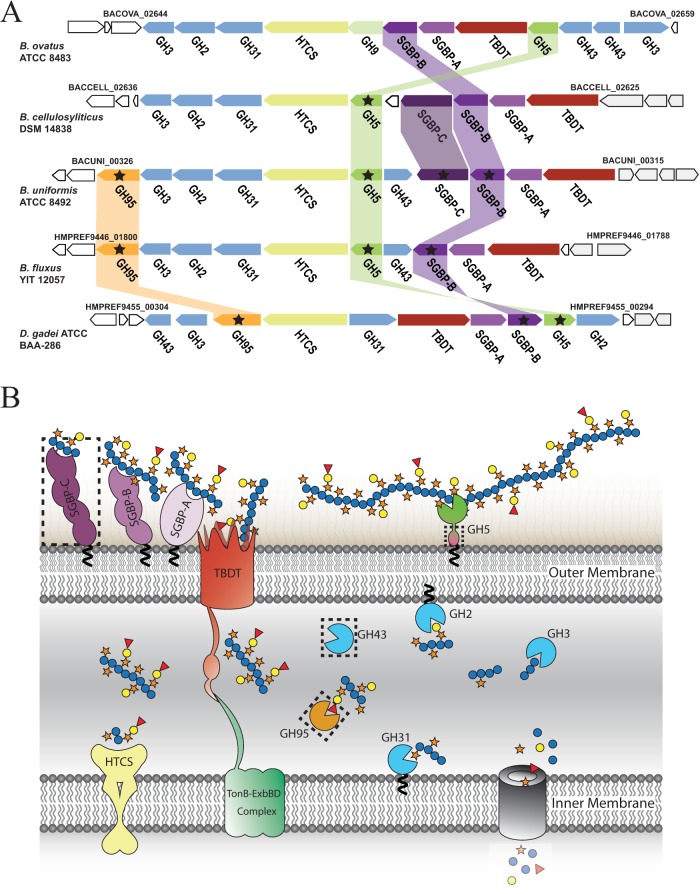
FIG 2.
XyGUL in Bacteroidetes. (A) Individual genes are represented as arrows indicating orientation, with genomic locus tags given for bounding genes. The encoded protein family is indicated below each gene: GHn, glycoside hydrolase family n member; SGBP, cell surface glycan-binding protein (SGBP-As are SusD homologs); HTCS, hybrid two-component system; TBDT, TonB-dependent transporter (SusC homologs). Homologous gene products studied in this work are connected by colored bars and are marked with stars. For clarity, homology between other genes/gene products (24) is not indicated. (B) Model of XyG saccharification in the cell envelope of Bacteroidetes. Gene products are colored as in panel A. SGBPs initiate polysaccharide recognition at the cell surface (38), where the GH5 (subfamily 4) endo-xyloglucanase cleaves the substrate (24) for active transport into the periplasm via the TBDT/SGBP-A complex (40). In the periplasm, exo-acting GHs work in a concerted fashion to liberate monosaccharides (24, 25) for cytosolic import and metabolism. The dashed rectangles indicate nonconserved protein products of the XyGUL. The model shows dicot fucogalactoxyloglucan utilization, which does not contain α-l-arabinofuranosyl residues that are the substrates of the GH43 members (24).