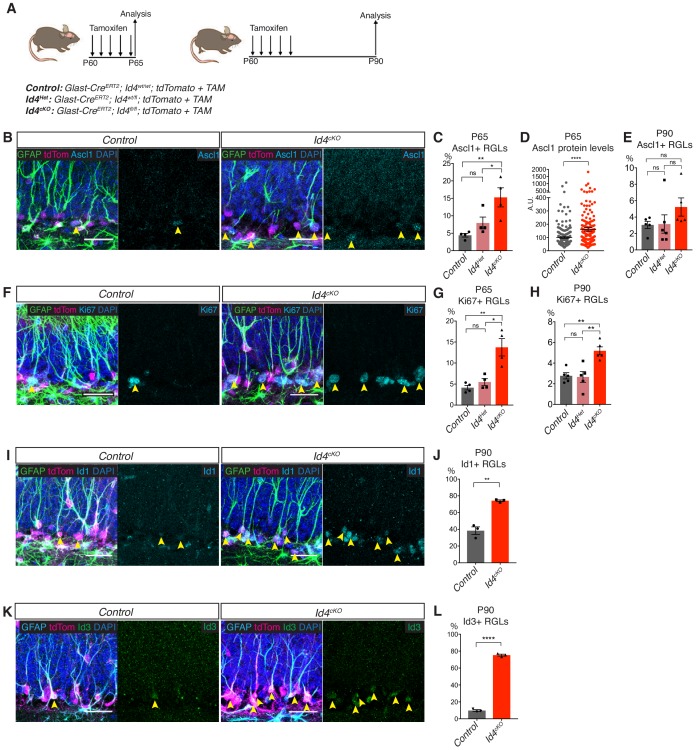
Figure 6. Loss of Id4 results in activation of quiescent RGLs in the adult hippocampus.
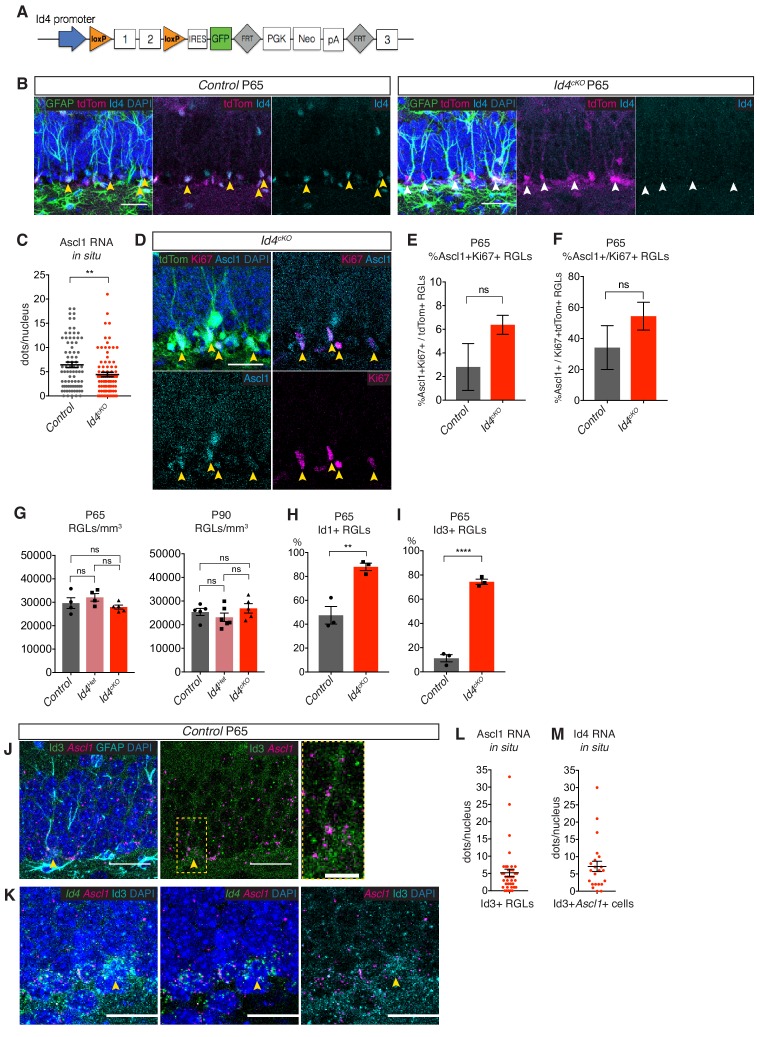
(A) Design of the experiment for acute and long-term deletion of Id4 from RGLs of the adult hippocampus using Id4cKO mice. (B) Immunolabeling for GFAP, tdTomato, Ascl1 and DAPI staining in control and Id4cKO mice after 5 days of tamoxifen administration. Yellow arrows indicate Ascl1-positive RGLs. Scale bar, 30 µm. (C–D) Quantification of Ascl1 protein in tdTomato+ RGLs in control, Id4Het and Id4cKO mice after 5 days of tamoxifen administration. Loss of both copies of Id4 results in increases in the number of Ascl1-expressing cells and in the levels of Ascl1 protein in RGLs (Control vs Het p=0.3276; Control vs cKO p=0.0067; Het vs cKO p=0.0381; protein levels p=2.01E-5). n = 4 for control, Id4Het and Id4cKO mice. (E) Quantification of Ascl1 protein in tdTomato+ RGLs control, Id4Het and Id4cKO mice 30 days after tamoxifen administration. The percentage of RGLs positive for Ascl1 is increased in Id4cKO mice compared with control mice 30 days after Id4 deletion (Control vs Het p=0.996; Control vs cKO p=0.311; Het vs cKO p=0.315). n = 5 for control and Id4Het mice, n = 6 for Id4cKO mice. (F) Immunolabeling for GFAP, tdTomato, Ki67 and DAPI staining in control and Id4cKO and control mice after 5 days of tamoxifen administration. Yellow arrows indicate Ki67-positive RGLs. Scale bar, 30 µm. (G–H) Quantification of the fraction of Ki67+ tdTomato+ RGLs in control, Id4Het and Id4cKO mice, 5 days (G) and 30 days (H) following tamoxifen administration. The percentage of Ki67+ tdTomato+ RGLs is strongly increased following acute deletion of both copies of the Id4 allele, and remained significantly increased, albeit to a lesser extent, following long-term deletion. (Control vs Het P65 p=0.7595, P90 p=0.980; Control vs cKO P65 p=0.0049, P90 p=0.0036; Het vs cKO p=0.0101, P90 p=0.0026). n = 4 for P65 control, Id4Het and Id4cKO mice at P65; n = 5 for P90 control, Id4Het and Id4cKO mice. (I) Immunolabeling for GFAP, tdTomato, Id1 and DAPI staining in control and Id4cKO and control mice 30 days after tamoxifen administration. Yellow arrows indicate Id1-positive RGLs. Scale bar, 30 µm. (J) Quantification of the fraction of Id1+ tdTomato+ RGLs 30 days after tamoxifen administration in control and Id4cKO mice. Loss of Id4 results in a 2-fold increase in the fraction of tdTomato+ RGLs positive for Id1 immunoreactivity, from 38.3 ± 4.5% to 74.1 ± 1.0% (p=0.0016). n = 3 for both control and Id4cKO. (K) Immunolabeling for GFAP, tdTomato, Id3 and DAPI staining in control and Id4cKO and control mice 30 days after tamoxifen administration. Yellow arrows indicate Id3-positive RGLs. Scale bar, 30 µm. (L) Quantification of the fraction of Id3+ tdTomato+ RGLs in (K). Id3 is increased by more than 8-fold in tdTomato+ RGLs following Id4 deletion, from 9.7 ± 1.0% to 75.3 ± 1.1% (p=1.87E-6). n = 4 for control mice and n = 3 for Id4cKO mice. Error bars represent mean ± SEM. Significance values: ns, p>0.05; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. See also Figure 6—figure supplement 1.