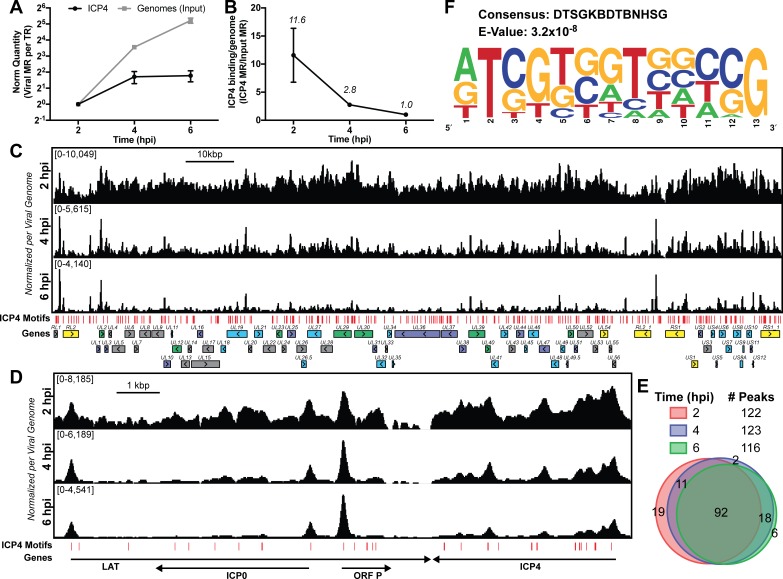
Figure 1. ICP4 binding at key points in the viral life cycle.
MRC5 cells were infected with HSV-1 for 2, 4, or 6 h, and ChIP-Seq for ICP4 was performed. (A) Quantification of viral genomes, measured as Input sample viral mapped reads (MR), normalized for sequencing depth or Total Reads (TR). Samples were normalized to two hpi, which was set as 1. (B) Quantification of ICP4 binding per viral genome measured as viral MR from ICP4 immunoprecipitation (IP) per viral MR from Input. (C–D) All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. Viral ORFs are indicated, color coded by gene class with IE as yellow, E as green, leaky late (L1) as blue, and true late (L2) as purple. Find Individual Motif Occurrences (FIMO) identified genome sequences matching the consensus motif in B are indicated in red. (E) Intersection of MACS2 identified ICP4 occupied regions. (F) ICP4 consensus binding motif.