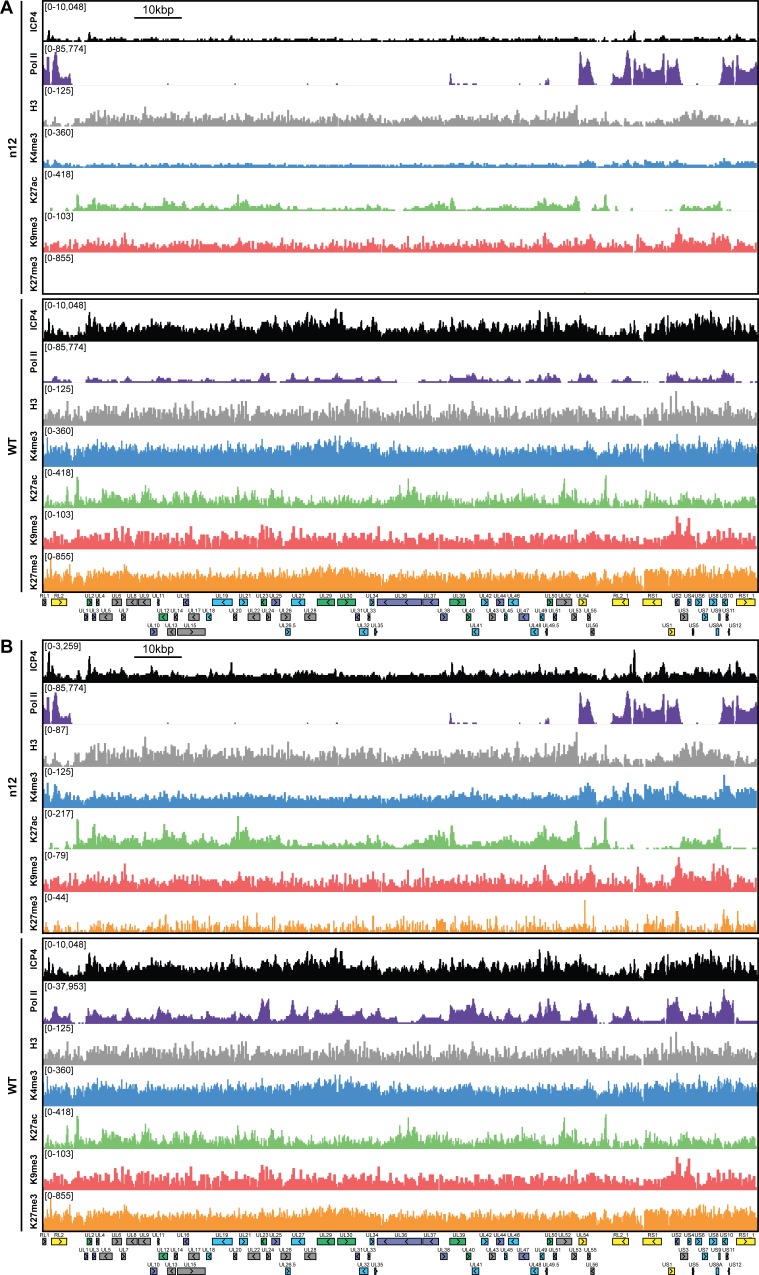
Figure 3. ICP4 dependence of viral genome accessibility and Histone H3 binding.
MRC5 cells were infected with an ICP4 null mutant (n12) or HSV-1 (WT) and harvested prior to genome replication. (A) ChIP-Seq for ICP4, Pol II, H3 and B–D) ATAC-Seq was performed. All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. (C) Quantitative analysis of ATAC-Seq data, measuring the relative tagmentation enrichment for the virus or host as compared to expected. (D) Histogram plot of ATAC-Seq fragment size for reads mapped to the viral (red) or cellular (black) genome. Mononucleosome protected fragments are approximately 180–250 bp.