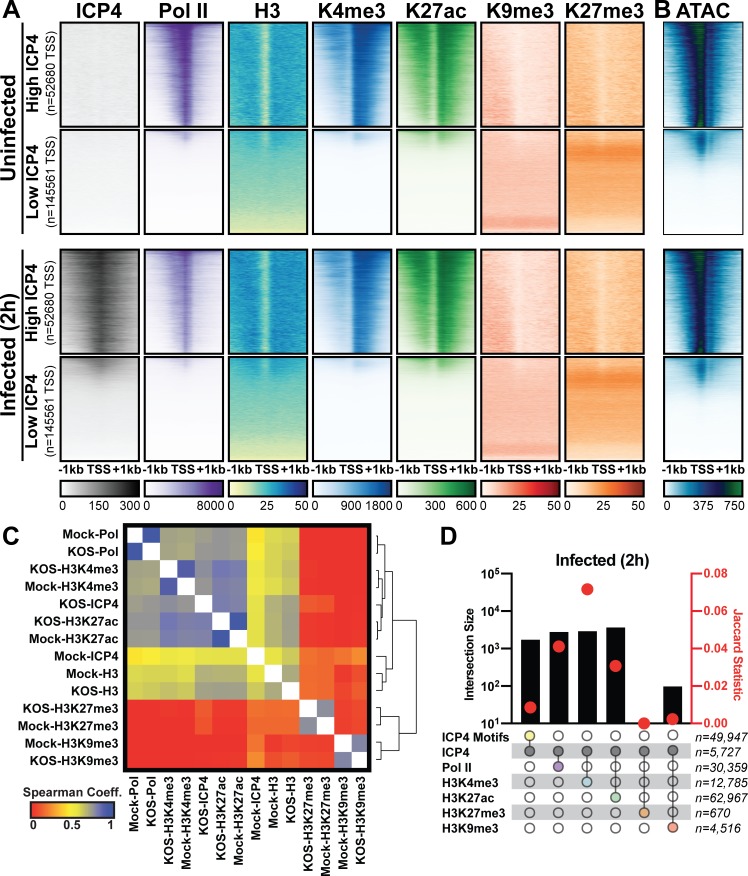
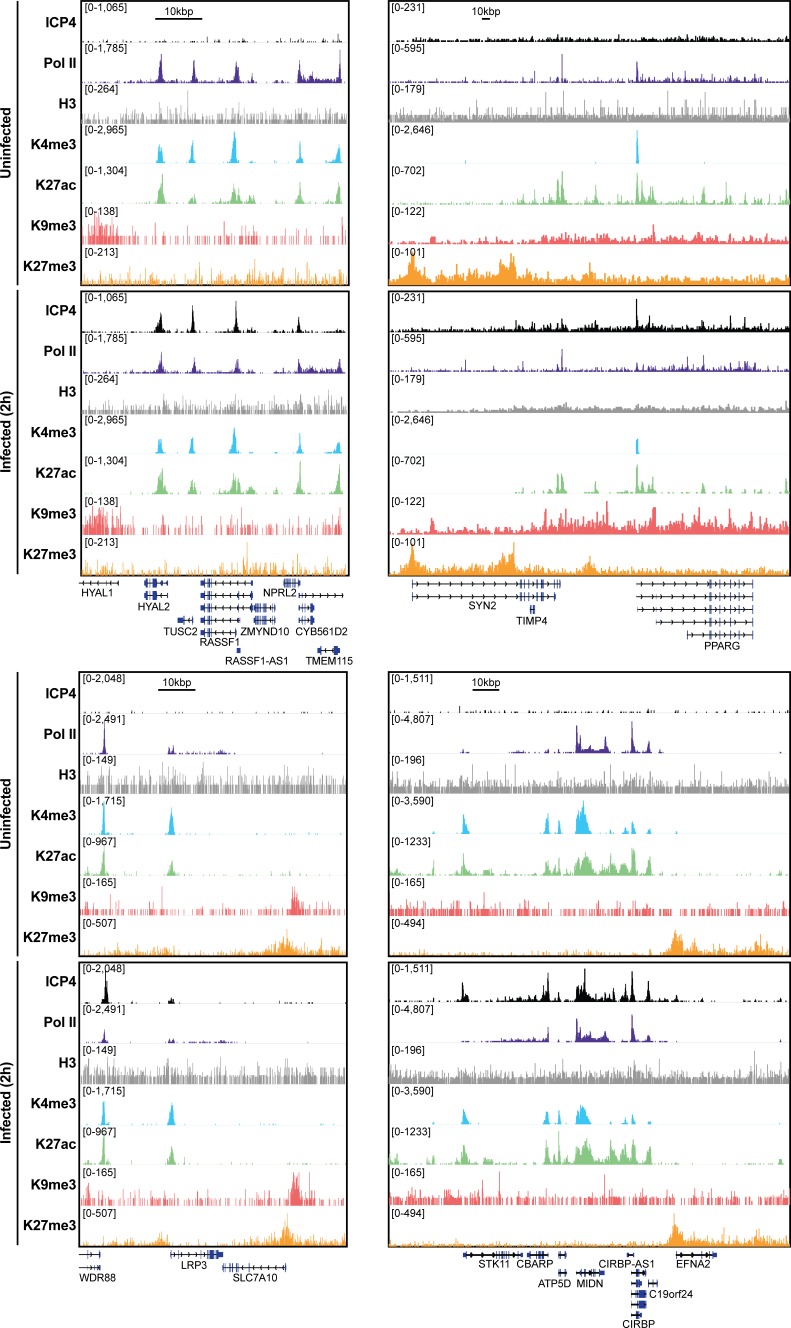
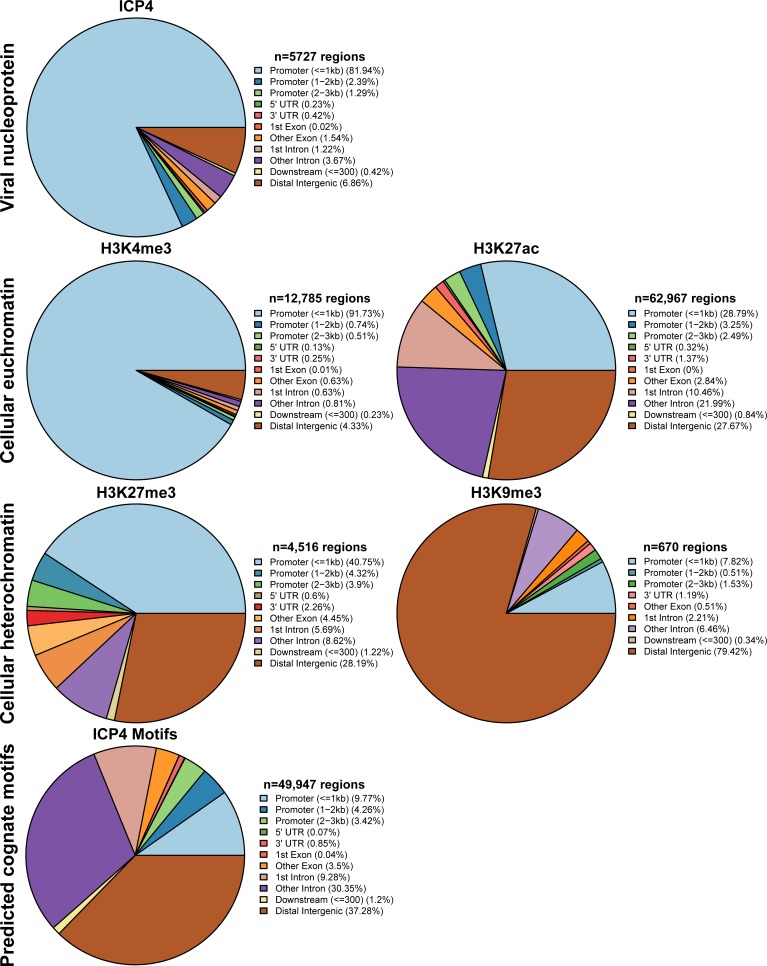
Figure 5. Association between cellular ICP4 binding and chromatin.
MRC5 cells were uninfected or infected with HSV-1 for 2 h. All data was aligned to the human genome (hg38) and normalized for sequencing depth. (A, C–D) ChIP-Seq data for ICP4, Pol II, H3, H3K4me3, H3K27acetyl, H3K9me3, and H3K27me3. (B) ATAC-Seq data. (A–B) Sequencing data centered + /- 1 kilobase from the TSS of cellular mRNAs. Data was stratified for ICP4 binding using K-means clustering. (C) Spearman correlation analysis, limited to cellular transcripts. (D) Intersection of MACS2 peaks, analyzed as number of intersecting peaks or Jaccard statistic.