Abstract
Satellite repeats are a structural component of centromeres and telomeres, and in some instances, their divergence is known to drive speciation. Due to their highly repetitive nature, satellite sequences have been understudied and underrepresented in genome assemblies. To investigate their turnover in great apes, we studied satellite repeats of unit sizes up to 50 bp in human, chimpanzee, bonobo, gorilla, and Sumatran and Bornean orangutans, using unassembled short and long sequencing reads. The density of satellite repeats, as identified from accurate short reads (Illumina), varied greatly among great ape genomes. These were dominated by a handful of abundant repeated motifs, frequently shared among species, which formed two groups: 1) the (AATGG)n repeat (critical for heat shock response) and its derivatives; and 2) subtelomeric 32-mers involved in telomeric metabolism. Using the densities of abundant repeats, individuals could be classified into species. However, clustering did not reproduce the accepted species phylogeny, suggesting rapid repeat evolution. Several abundant repeats were enriched in males versus females; using Y chromosome assemblies or Fluorescent In Situ Hybridization, we validated their location on the Y. Finally, applying a novel computational tool, we identified many satellite repeats completely embedded within long Oxford Nanopore and Pacific Biosciences reads. Such repeats were up to 59 kb in length and consisted of perfect repeats interspersed with other similar sequences. Our results based on sequencing reads generated with three different technologies provide the first detailed characterization of great ape satellite repeats, and open new avenues for exploring their functions.
Keywords: heterochromatin, satellite repeats, long sequencing reads, great apes
Introduction
Heterochromatin is the gene-poor and highly compacted portion of the genome. It is typically dominated by satellite repeats—long arrays of tandemly repeated noncoding DNA (Kit 1961; Sueoka 1961) that consist of smaller units organized into higher order repeat structures. Heterochromatin is abundant, for instance, at telomeres and centromeres of human chromosomes (Sujiwattanarat et al. 2015). While labeled as “junk DNA” in the past, heterochromatin was later found to fulfill important functions in the genome (Walker 1971; Yunis and Yasmineh 1971; Ferree and Barbash 2009). Heterochromatin satellite repeat expansions have been associated with changes in gene expression and methylation (Brahmachary et al. 2014; Quilez et al. 2016). It has also been proposed that heterochromatin aids in maintaining cellular identity by repressing genes that are not specific to a particular cell lineage (reviewed in Becker et al. 2016). For instance, the heterochromatin-associated histone mark H3K9me3 blocks reprogramming to pluripotency (Soufi et al. 2012). Additionally, heterochromatin loss is part of the normal aging process (Zhang et al. 2015) and changes during stress (Gowen and Gay 1933; Jolly et al. 2004; Rizzi et al. 2004; Tittel-Elmer et al. 2010; Seong et al. 2011). Despite a growing interest in understanding these important functions of heterochromatin, satellite repeats are frequently underrepresented in genomic studies—due to the difficulties in sequencing and assembling these highly similar sequences (Chaisson et al. 2015). The lack of information about satellite repeats is particularly alarming given their high abundance; for example, alpha satellites were estimated to constitute ∼3% of the human genome (Manuelidis 1978; Hayden et al. 2013). Relatedly, satellite repeats are likely plentiful in yet unassembled gaps in the human genome (Miga et al. 2014; Stephens and Iyer 2018). One of the largest uncharacterized gaps in the human genome is located in the Male-Specific region of the Y chromosome (MSY), which contains six types of satellite repeat sequences (DYZ1, DYZ2, DYZ3, DYZ17, DYZ18, and DYZ19) (Skaletsky et al. 2003).
Heterochromatin exhibits remarkable interspecific variability in size and structure. Such variability can be frequently observed even between closely related species. For instance, on the long arm of the Y chromosome, heterochromatin is the major component in human and gorilla, but is virtually absent in chimpanzee (Gläser et al. 1998)—notwithstanding the fact that human, gorilla, and chimpanzee diverged <8 million years (My) ago (Glazko and Nei 2003). As another example, whereas 20% of the genome of Drosophila melanogaster is composed of satellite DNA, this percentage is as low as 0.5% for D. erecta and as high as 50% for D. virilis (Gall et al. 1971; Lohe and Brutlag 1987); the estimated divergence time between D. erecta and D. melanogaster is 13 My, and is 63 My between D. virilis and D. melanogaster (Tamura et al. 2004). The differences in satellite repeat abundance in nine Drosophila species were proposed to result predominantly from lineage-specific gains accumulated over the past 40 My of evolution (Wei et al. 2018). Due to its rapid evolutionary turnover, heterochromatin can serve as a species barrier (Yunis and Yasmineh 1971; Ferree and Barbash 2009; Rošić et al. 2014).
Profound intraspecific variability in heterochromatin has also been reported, including that among humans (Altemose et al. 2014; Miga et al. 2014). For instance, the length of the DYZ1 satellite repeat varies considerably among major Y chromosome haplogroups; DYZ1 is longer in Y chromosomes belonging to the predominantly Asian O haplogroup than in those belonging to the predominantly African E haplogroup (Altemose et al. 2014). The centromeric array of the X chromosome was shown to vary in length among different human populations by as much as an order of magnitude (0.5–5 Mb) (Miga et al. 2014). Some human neocentromeres were found to harbor only very short (as short as 15 kb) heterochromatin domains leading to a defect in sister chromatid cohesion (Alonso et al. 2010).
In addition to satellite repeats with relatively long repeat units (e.g., alpha satellites with repeat unit of ∼171 bp), three classes of satellite repeats with unit sizes ≤50 bp are of particular interest due to their abundance or function in great apes. These include (AATGG)n satellite, telomeric satellite (TTAGGG)n, and AT-rich 32-unit subterminal satellites (StSats). The (AATGG)n repeat is the source of Human Satellites 2 and 3 (HSat2 and HSat3) (Altemose et al. 2014). On chromosome 9, it also encodes a long noncoding RNA that is critical for the heat shock response in human cells (Goenka et al. 2016). Previous studies investigated the variability, abundance, and length distribution of the (AATGG)n repeat in the human genome (Tagarro et al. 1994; Skaletsky et al. 2003; Subramanian et al. 2003; Altemose et al. 2014). This repeat was also identified in orangutan, chicken, maize, sea urchin, and Daphnia (Grady et al. 1992; Flynn et al. 2017), however, its variation in great ape species was never studied. The telomeric (TTAGGG)n satellite functions to maintain genome stability; telomere loss is correlated with cell division and aging (Lanza et al. 2000; Rizvi et al. 2014). StSats present in the genomes of chimpanzee, bonobo, and gorilla (Royle et al. 1994) localize proximal to telomeres (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012) and were proposed to play a role in telomere metabolism (Novo et al. 2013) and meiotic telomere clustering important for homolog recognition and pairing (in a process similar to that identified in plants; Bass et al. 2000; Calderón et al. 2014).
In this study, we characterize turnover of satellites with repeat units ≤50 bp among six great ape species—human, chimpanzee, bonobo, gorilla, Bornean orangutan, and Sumatran orangutan—which diverged <15 My ago (Goodman et al. 2005). We focus on repeats that constitute portions of long arrays of satellite DNA and use them as a proxy for heterochromatin (Wei et al. 2014). This approximation is needed because of challenges in the direct identification of heterochromatin due to its transient nature in various cells of individuals throughout their lifetime. In this manuscript, we, first, identify satellite repeats in short sequencing reads generated with the low-error rate Illumina technology, and investigate their inter- and intraspecific variation. We pinpoint repeats with higher incidence in males than females and, for some of these repeats, confirm location on the Y chromosome using existing Y assemblies or fluorescent in situ hybridization (FISH). Next, we use the repeated motifs identified from low-error rate short reads as queries to decipher the lengths and densities of ape satellite repeats from error-prone long reads (both Pacific Biosciences, or PacBio, and Oxford Nanopore, or Nanopore). To the best of our knowledge, ours is the first study of inter- and intraspecific satellite repeat variability, repeat expansions and correlations, as well as of male-biased repeats, in great apes.
Results
Repeat Identification in Short Reads
To study inter- and intraspecific variability of satellite repeats in great apes, we utilized 100- or 150-bp Illumina sequencing reads generated for 79 individuals (57 females and 22 males; supplementary table S1, Supplementary Material online) as a part of the Ape Diversity Project (ADP) (Prado-Martinez et al. 2013). These included chimpanzees (Nigeria-Cameroon, Eastern, Central, and Western chimpanzees), bonobos, gorillas (Eastern lowland, Cross river, and Western lowland gorillas), Sumatran orangutans, and Bornean orangutans (supplementary table S1, Supplementary Material online). Additionally, in order to match the library preparation protocol that was used for these great ape data, we used sequencing reads for 9 human males from diverse populations generated as part of the Human Genome Diversity Project (HGDP) (Cann et al. 2002; Rosenberg et al. 2002; Meyer et al. 2012). After filtering (see Materials and Methods), in this set of 79 + 9 = 88 individuals, the median number of reads per individual was 190,722,592 (supplementary table S1, Supplementary Material online).
Sequencing reads are expected to present a more complete picture of satellite repeat distributions than the existing reference genome assemblies (Lower et al. 2018). To annotate repeats in sequencing reads, we used Tandem Repeats Finder, TRF (Benson 1999) (when available, 150 bp reads were trimmed to 100 bp for consistency) and focused on repeats with a repeated unit of ≤50 bp (so that at least two units could fit within a 100 bp read). This approach does not allow detection of satellites with longer repeated units, such as centromeric alpha satellites, for which even a single repeated unit would not fit within a short sequencing read, however is geared toward accurate identification of satellite repeats with shorter repeated units. Additionally, in order to study long satellite arrays likely to be present in the heterochromatin, we only retained sequencing reads in which repeated arrays covered at least 75% of the read length (i.e. ≥75 bp, see Materials and Methods). This effectively removed most microsatellites from our data set. As a result, we identified 5,494 distinct repeated motifs (later called satellite repeated motifs, or repeated motifs) across the studied species and verified that they were not artifacts of read length or software choice (supplementary note 1, Supplementary Material online).
Inter- and Intraspecific Variability
Repeat Density Varies among Great Ape Species
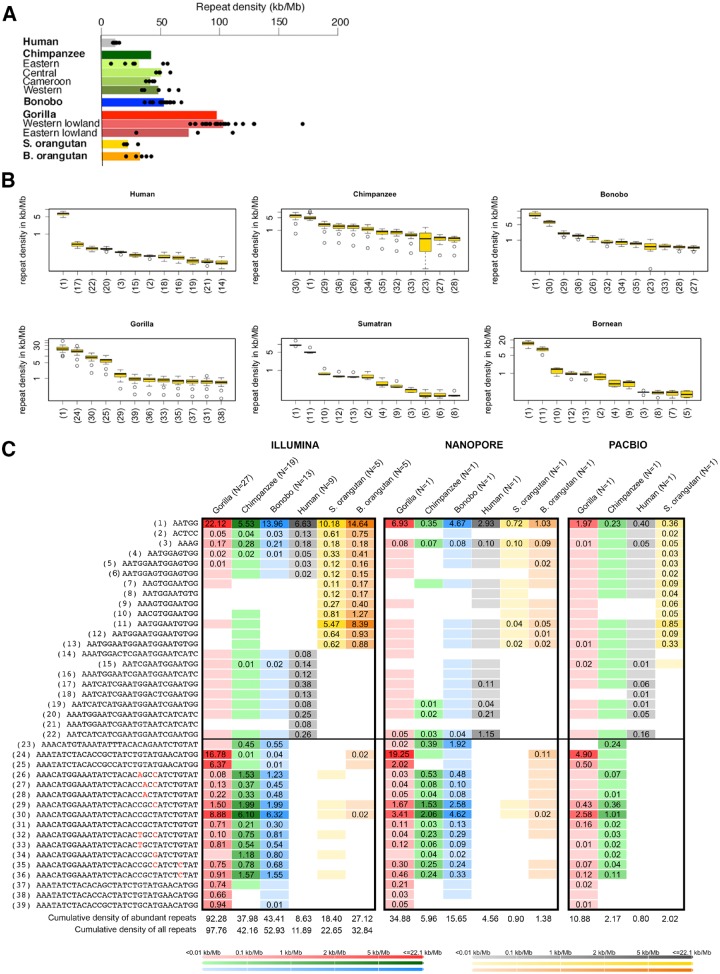
We compared the overall satellite repeat density (computed cumulating occurrences for all types of repeated motifs) among the studied ape species and subspecies (fig. 1A). For each individual, satellite repeat density (later called repeat density) was computed as the total number of kilobases annotated in satellite repeats per million bases of sequencing reads (kb/Mb). First, we verified that technical replicates—different Illumina lanes/runs for the same individual—had highly correlated repeat densities (supplementary fig. S1, Supplementary Material online). Second, we verified that repeat density and sequencing depth were not correlated with each other (supplementary fig. S2, Supplementary Material online). Illumina PCR+ libraries were generated for ADP (Prado-Martinez et al. 2013) and HGDP (Cann et al. 2002; Rosenberg et al. 2002; Meyer et al. 2012); while the types of repeated motifs identified were likely unaffected by the amplification step during library preparation, their densities might have been (supplementary note 2, Supplementary Material online), and thus the precise repeat densities we report here might differ from the actual densities in the studied genomes. However, biases due to PCR amplification should be limited (see next paragraph for an analysis of human PCR− libraries suggesting minimal bias). Moreover, because all samples were processed with the same library preparation protocol, any existing biases should be concordant and should not affect comparisons of numbers among and within species (fig. 1A). We observed the highest average repeat densities (across individuals) in Western and Eastern lowland gorillas (103 and 74.0 kb/Mb, respectively), and the lowest in human (11.9 kb/Mb) and Sumatran orangutan (22.6 kb/Mb).
Fig. 1.
Densities and similarity among satellite repeats in great apes. (A) Intra- and interspecific variation in overall repeat density. Repeat densities are plotted for each species and subspecies. Each dot represents a single individual, and bars are mean values. For species comprising subspecies, a species-level average is also represented with a bar. Human (N = 9, black), bonobo (N = 13, blue), chimpanzee (N = 19, green), gorilla (N = 27, red), S. orangutan is Sumatran orangutan (N = 5, yellow), and B. orangutan is Bornean orangutan (N = 5, orange). The cross river gorilla has sample size of 1 and is only included in the species-level analysis. (B) Boxplots of repeat densities are plotted for each species showing intraspecies variability for the most abundant repeated motifs in each species. The repeated motifs are numbered as in figure 1C. Ticks on the y axis represent 1, 5, 10, 20, 30, 40, and 50 kb/Mb. Plotted with OUTLINE=FALSE to remove outliers. (C) Heatmap of average repeat densities (across individuals) for the 39 abundant repeats in each of the six species. Color coding from dark to light blue represents high to low values. Repeats present at <100 loci per 20 million Illumina reads or not found with NCRF in long-read data are considered absent (white cells). Repeats with densities <0.01 kb/Mb are only present in trace amounts (blank colored cells). (AATGG)n-derived and 32-mer-derived repeated motifs are separated by a horizontal line. Cumulative densities of abundant repeats and of all repeats are calculated as averages across all individuals.
Great Ape Genomes Harbor Only a Handful of Abundant Repeated Motifs, Many of Which Are Shared among Species and Are Phylogenetically Related
We next investigated whether great ape genomes possess a few highly abundant repeated motifs, or many different repeated motifs present at relatively low abundance. We ranked motifs by descending abundance and found that the six great ape species we considered (subspecies were combined for this analysis) contain only a small number of abundant repeated motifs: usually ≤12 in each of the species (supplementary fig. S3, Supplementary Material online). There were a total of 39 unique motifs with density ranking 12 or higher in the six species analyzed (supplementary fig. S3 and table S2, Supplementary Material online and fig. 1B). These 39 repeated motifs had overall average densities (across individuals) of 8.63, 38.0, 43.4, 92.3, 18.4, and 27.1 kb/Mb in the six species (fig. 1C), and represent ∼73%, 90%, 82%, 94%, 81%, and 83% (i.e., very large portions) of the total satellite repeat density we found in the human, chimpanzee, bonobo, gorilla, Sumatran orangutan, and Bornean orangutan genomes, respectively. Notably, when we compared densities of these 39 repeats between nine humans sequenced with the PCR+ protocol used also for nonhuman apes throughout our study and nine other humans sequenced with a PCR− protocol (supplementary fig. S4, Supplementary Material online), we observed minimal differences beyond expected interindividual variation, suggesting only small effects of PCR amplification on our repeat density estimates.
Additionally, we searched for the (TTAGGG)n repeat that is important for telomere protection and which we expected to be present in our data set, yet we did not find it among the most abundant motifs discussed in the previous paragraph; in our data, this repeat has ranks 42, 112, 144, 321, 43, and 38 in the genomes of human, chimpanzee, bonobo, gorilla, Sumatran orangutan, and Bornean orangutan genomes, respectively (supplementary fig. S3, Supplementary Material online). It constitutes ∼0.23%, 0.10%, 0.06%, 0.02%, 0.10%, and 0.11% of the total satellite repeat density in each of these genomes, with repeat density ranging from 0.0227 to 0.0422 kb/Mb among species (supplementary table S3, Supplementary Material online).
The 39 abundant repeated motifs we identified had varying levels of sharing among species (fig. 1C). Six motifs were present in all six species analyzed. The (AATGG)n repeat, shared by all six species, was the most abundant repeat in humans (with an average density of 6.63 kb/Mb) as well as in gorilla, bonobo, Sumatran orangutan, and Bornean orangutan (with average densities of 22.1, 14.0, 10.2 and 14.6 kb/Mb, respectively), and the second most abundant repeat in chimpanzee (with an average density of 5.53 kb/Mb). The next most abundant repeated motifs in human and orangutans were phylogenetically related to the (AATGG)n (fig. 1C;supplementary fig. S5 and table S2, Supplementary Material online). Their overall average densities (excluding (AATGG)n itself) were 1.75, 9.22, and 13.7 kb/Mb in the genomes of human, Sumatran orangutan, and Bornean orangutan, respectively (fig. 1C). In addition to (AATGG)n and repeated motifs related to it, we identified highly similar StSat 32-mers (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012) and a 31-mer related to them (all differing by 1–2 bases; fig. 1C and supplementary fig. S5, Supplementary Material online). These repeats were abundant in the genomes of chimpanzee, bonobo, and gorilla with overall average densities of 15.8, 15.8, and 39.6 kb/Mb, respectively. In fact, one of these 32-mers was the motif with the highest repeat density in chimpanzee (6.10 kb/Mb). 32-mers were absent from the human genomes analyzed, and were very sparse in the orangutan genomes (fig. 1C). We found no relationship between the degree to which a repeated motif was shared across the six species and its repeat density (supplementary fig. S6, Supplementary Material online). In conclusion, the overall satellite repeat content in great ape genomes appears to be driven by only a few highly abundant repeated motifs, many of which are shared among species and are phylogenetically related to each other (supplementary fig. S5, Supplementary Material online).
The Majority of Less-Abundant Repeated Motifs Are Species-Specific
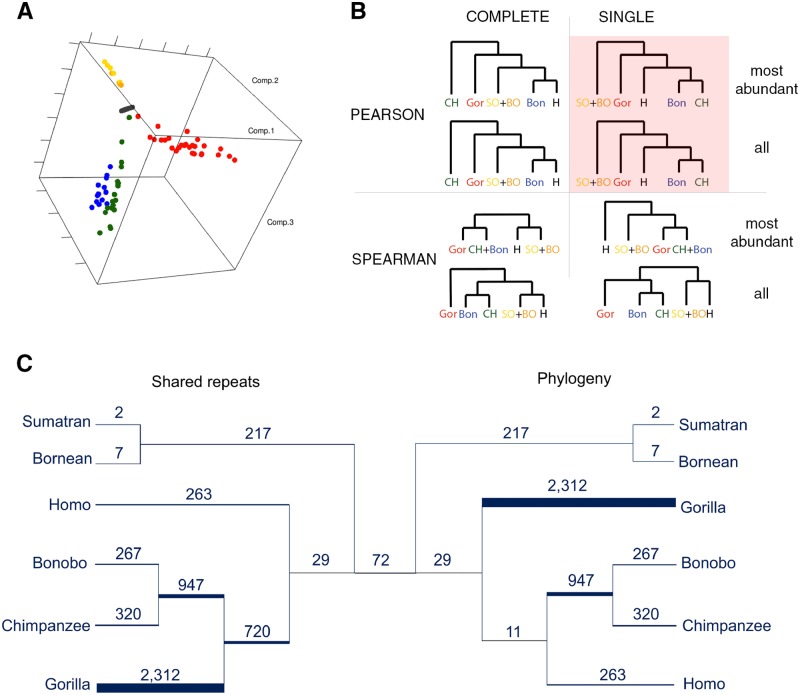
We subsequently analyzed the 5,455 repeated motifs constituted by the initial set minus the 39 abundant repeats discussed in the previous section, and found substantial differences among great ape species when profiling their presence/absence (supplementary fig. S7A, Supplementary Material online). Despite the relatively recent divergence of the species considered (Goodman et al. 2005), as many as 3,170 of the 5,455 distinct repeated motifs were species-specific. Among them, 2,312 were gorilla-specific, while only 262 were human-specific. As expected, the chimpanzee and bonobo sister species shared many repeated motifs (a total of 947, representing 75% and 78% of all repeats identified in each species, respectively), and so did the Sumatran and Bornean orangutan sister species (a total of 217, representing 99% and 97% of all repeats identified in each species, respectively). Interestingly, we found a positive relationship between the number of species-specific repeated motifs and mean repeat density in a species (supplementary fig. S8, Supplementary Material online; human is an outlier in this analysis). These results did not change qualitatively when we considered the same number of individuals per species (supplementary figs. S7B–G and S8B, Supplementary Material online).
The majority of the 39 abundant motifs were present in all individuals of a given species (supplementary fig. S7H, Supplementary Material online) but exhibited substantial variability in repeat density among them (supplementary fig. S9, Supplementary Material online). For instance, the average fold difference for the (AATGG)n repeat among two unrelated human males in our study was 1.23 (supplementary table S4, Supplementary Material online). Other motifs, especially those of lower abundance, although identified in a species, were only present in a subset of individuals (supplementary fig. S7A, Supplementary Material online).
Relatedness of the Studied Species Based on Satellite Repeat Data
We investigated whether the satellite content alone was sufficient to differentiate among species and to recapitulate the accepted species phylogeny. We found that, first, individuals from the same species generally clustered together. Using principal component analysis (fig. 2A and supplementary note 3, Supplementary Material online) and repeat densities, individuals belonging to different species formed fairly well-separated groups (with first three principal components explaining 98% of the variance). Abundant repeated motifs were sufficient for species classification: individuals were assigned to species with ∼96% accuracy with a Linear Discriminant Analysis and ∼92% accuracy with a Random Forest classifier (supplementary note 3 and table S5, Supplementary Material online; the list of repeated motifs most discriminating among species is provided in supplementary note 3, Supplementary Material online). Second, we found that many repeated motifs are shared among species in a manner inconsistent with the accepted species phylogeny. After hierarchical clustering of all individuals based on repeat densities, different ways of clustering and different distance metrics resulted in variations in the tree topology (supplementary note 3, Supplementary Material online), with most of the topologies being incompatible with the accepted phylogeny (fig. 2B). In fact, the higher level agglomeration only reproduced the accepted species phylogeny in scenarios with Pearson correlations and single linkage function (fig. 2B and supplementary fig. S10B, D, Supplementary Material online). Moreover, we occasionally (e.g., when using only 39 repeated motifs) noticed intermixing between the two orangutan species and between chimpanzee and bonobo. We observed a similar pattern estimating a phylogeny based solely on the number of shared repeated motifs (in terms of their presence/absence). Chimpanzee, bonobo, and gorilla (sharing 720 repeated motifs) formed a cluster that did not include human (fig. 2C, left), departing from the accepted great ape species phylogeny (fig. 2C, right). This result did not change after we excluded StSat 32-mers (supplementary fig. S11, Supplementary Material online). Taken together, both distances in repeat densities (fig. 2B) and configurations of shared (vs. not shared) repeats (fig. 2C) across species, show a distortion of the signals as compared with the accepted species phylogeny. This suggests an especially rapid evolution of satellite repeats among great apes.
Fig. 2.
Relatedness of 88 analyzed individuals belonging to the six great ape species. (A) Principal Component Analysis. Individuals are plotted as circles in the space of the first three principal components extracted from the densities of the 39 abundant repeats, which explain 98% of the variance. Colors correspond to the six species: human (black), bonobo (blue), chimpanzee (green), gorilla (red), Sumatran orangutan (orange), and Bornean orangutan (gold). (B) Hierarchical clustering of individuals, using combinations of Pearson or Spearman correlation coefficients, single or complete linkage, and most abundant (N = 39) or all motifs (N = 5,494). The topology agreeing with the accepted species phylogeny is shown on the pink background. (C) Species topology based on repeats presence/absence. A schematic figure showing repeated motifs unique to a species (terminal branches) and those that are shared among the species descending from internal branches. On the left, the tree is built based on the presence/absence of repeated motifs, iteratively joining species sharing the most repeated motifs. On the right, the tree is built according to the accepted species phylogeny (Goodman et al. 2005) and the number of shared repeated motifs is indicated. The branch widths are proportional to the number of repeated motifs (branch lengths are uninformative). About 72 repeated motifs (the number shown in the middle) were shared among all six studied species.
Male-Biased Repeats
Male-Biased Repeats Are among the Most Abundant
We next focused on identifying repeats potentially located on great apes Y chromosomes, based on the expectation that they should be substantially more frequent in males than in females, that is, male-biased. We considered all chimpanzee, bonobo, and gorilla individuals, as well as ten orangutan individuals (combining five Sumatran and five Bornean). In addition to the nine human males from HGDP (Cann et al. 2002; Rosenberg et al. 2002; Meyer et al. 2012), we also used three fathers and three mothers from human trios (supplementary table S1, Supplementary Material online and see Materials and Methods). We restricted attention to repeated motifs with density >0.5 kb/Mb in any given species. For each such motif, we calculated the average male-to-female density ratio across individuals and assessed significance of the difference in repeat density between males and females with a Mann–Whitney test (supplementary table S6, Supplementary Material online). Our analysis resulted in a total of 18 male-biased repeated motifs (P < 0.2), which are candidates to be located on great apes Y chromosomes: 1 in human ((AATGG)n), 5 in chimpanzee, 9 in bonobo, 14 in gorilla, and 1 in orangutans ((ACTCC)n) (supplementary table S6, Supplementary Material online). Interestingly, all the male-biased motifs (P < 0.2) were among the most abundant repeated motifs in the ape genomes (ranging between 1st and 14th in the species-specific ranks).
Male-Biased 32-Mers Can Be Found on the Gorilla and Bonobo Y Chromosomes
We further restricted attention to male-biased 32-mer StSats, which had higher incidence in males than females in chimpanzee, bonobo, and gorilla (supplementary table S6, Supplementary Material online), and searched for additional evidence that they indeed might be located on the Y chromosomes of these species. First, we screened the Y chromosome assemblies of chimpanzee (Hughes et al. 2010) and gorilla (Tomaszkiewicz et al. 2016) for occurrences of these male-biased StSats (see Materials and Methods; no bonobo Y chromosome assembly is currently available). We found them in the latter but not in the former. This could be explained by the fact that long PacBio reads, which are more likely to capture these StSats, were used to generate the gorilla’s Y assembly, and not the chimpanzee’s. However, it is also possible that some of these StSats are indeed absent from the chimpanzee Y chromosome (see next paragraph).
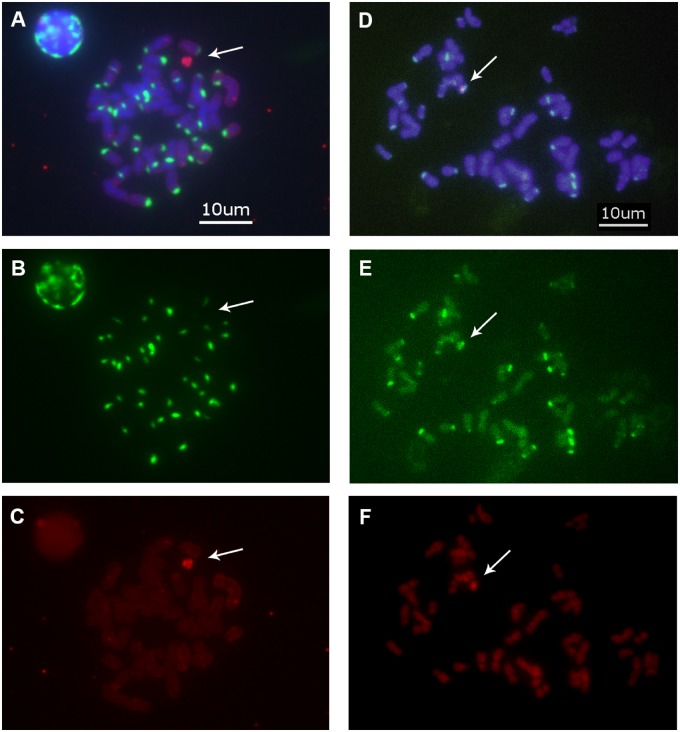
Second, to experimentally assess whether male-biased StSats (supplementary table S6, Supplementary Material online) are present on the Y chromosomes of bonobo and chimpanzee, we performed FISH. We used two probes (see Materials and Methods); the degenerate Pan32 probe containing the sequences of two male-biased StSats (supplementary table S6, Supplementary Material online), and the whole bonobo Y chromosome (WBY) probe containing the flow-sorted bonobo Y chromosome. These probes were hybridized to metaphase spreads of bonobo and chimpanzee males. The StSat probe hybridized to (sub)telomeric locations of most chromosomes (fig. 3A and B and D and E), suggesting an association with heterochromatin. Moreover, both probes hybridized to the bonobo Y chromosome, confirming Y localization (fig. 3A)—consistent with our computational predictions (the P values for bonobo male-to-female abundance differences were 0.03 and 0.05 for the two StSats included in the degenerate probe; supplementary table S6, Supplementary Material online). FISH could not confirm the presence of the same StSat probe on the chimpanzee Y chromosome (fig. 3D)—again consistent with our computational analysis, which provided only weak evidence of male bias for the studied StSats in chimpanzee (P values of 0.2 and 0.2 for the two StSats included in the degenerate probe; supplementary table S6, Supplementary Material online). In summary, we identified several male-biased repeats in the genomes of great ape species, and for a number of them, we were able to validate their Y chromosome location either by examining Y assemblies or by FISH experiments.
Fig. 3.
Fluorescent in situ hybridization (FISH) analysis. Hybridization of both the WBY and Pan32 probes to DAPI-counterstained chimpanzee male chromosomes (A) and bonobo male chromosomes (D), of only the Pan32 probe to DAPI-counterstained chimpanzee male chromosomes (B) and bonobo male chromosomes (E), of only the WBY probe to DAPI-counterstained chimpanzee male chromosomes (C) and bonobo male chromosomes (F). The bonobo Y chromosome is positive for both probes, whereas the chimpanzee Y chromosome is positive only for the WBY probe and not for the Pan32 probe. The Pan32 probe: 5′-amine-modified oligonucleotide probe with a candidate 32-mer male-biased motif sequence is labeled with Alexa Fluor (green). The WBY probe: whole bonobo Y chromosome painting probe is labeled with digoxigenin (red). The white arrows indicate the location of the Y chromosome. Scale bar = 10 μm.
Estimating Satellite Repeat Abundance and Length with Long-Read Data
Because short-read technologies can only provide information about total repeat abundances, and satellite repeats are routinely underrepresented in sequenced assemblies, one can take advantage of long reads, for example, as produced by Nanopore or PacBio, to provide a presumably less-biased view of repeated array lengths. However, there could still be technology-specific differences in repeat densities because of potential biases in each technology. Prior to analyzing the lengths of repeat arrays in long Nanopore and PacBio reads, we assessed the extent to which the repeat densities from these long-read technologies corresponded to those obtained with Illumina. We studied a familial trio from the Genome in a Bottle Consortium (Zook et al. 2016) that was sequenced using all three technologies. This analysis revealed that the use of the sequencing technology has an effect on the repeat density, although we still could consistently differentiate between lowly and highly abundant motifs independent of the technology used (supplementary note 4, Supplementary Material online and fig. 1C).
Unfortunately, adequate software to retrieve repeats from long sequencing reads, which are notoriously error-prone (with error rates around ∼15–16% for both PacBio and Nanopore; Rhoads and Au 2015; Jain, Koren, et al. 2018), does not currently exist. To address this limitation, we developed Noise-Cancelling Repeat Finder (NCRF; Harris et al. 2019), a stand-alone software that can recover repeat length distributions from long reads notwithstanding their high error rates. Briefly, NCRF aligns a user-specified motif to a DNA fragment in a read, with a motif looped as often as needed. It uses a Dynamic Programming matrix, organizing each nucleotide of a motif in a row and wrapping a DNA sequence in columns, allowing for looping from the end of the motif to the beginning. Additionally, NCRF employs technology-specific scoring parameters and affine gap penalties. Initially, NCRF identifies continuous arrays of highly similar repeated motifs (imperfect repeats). This is vital as arrays comprising a dominant motif and one or more derived motifs represent an important facet of biological variability (Plohl et al. 2008). Since the direct de novo identification of satellite repeats from error-prone long reads is challenging, we used the 39 abundant Illumina-derived repeated motifs identified above (see Repeat Identification in Short Reads) as queries for the screening of long reads by NCRF.
To evaluate densities and lengths of these 39 motifs in long-read technologies using NCRF, we sequenced six great ape individuals, one from each species of great apes, on one Nanopore MinION flow cell (supplementary tables S7 and S8, Supplementary Material online), and employed publicly available PacBio sequencing reads available for four great ape species (supplementary tables S7 and S9, Supplementary Material online) (Gordon et al. 2016; Kronenberg et al. 2018). For our Nanopore data, the longest observed read was 206 kb and the read length N50 ranged from 26 to 37 kb among samples (supplementary table S8, Supplementary Material online). In comparison, using a single flow cell of publicly available PacBio data for each species, the longest observed read was 184 kb and the read length N50 ranged from 19 to 34 kb among samples (supplementary table S9, Supplementary Material online). Concerning repeat densities we found with NCRF, for both PacBio and Nanopore reads, the general patterns were consistent with those inferred from Illumina reads with TRF (fig. 1C)—however, the exact densities differed. The differences can be in part explained by the fact that different individuals of the same species were sequenced using each technology. An additional factor could be that Nanopore and PacBio reads employ distinct library preparation and sequencing protocols that are prone to different biases (see Discussion and supplementary note 4, Supplementary Material online). Interestingly, some of the repeated motifs abundant in short-read data, such as the (AATGG)n repeat, were not as abundant in long-read data.
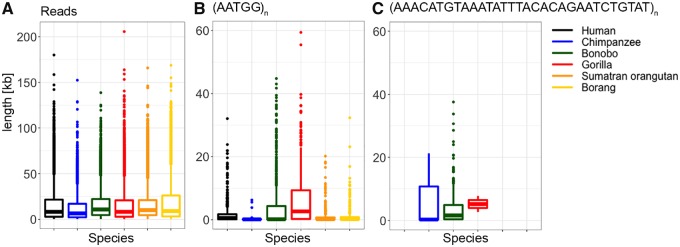
We also discovered that long satellite arrays were frequently a mix of more than one motif, present in perfect patches interspersed with highly similar, yet different, sequences. To come to this conclusion, we proceeded as follows. First, we verified that long reads were able to capture the full lengths of satellite repeats (supplementary figs. S12 and S13, Supplementary Material online), as demonstrated by the fact that in the majority of cases long reads encompassed complete repeat arrays (depending on the species, 90–95% and 99% of repeat arrays were nested within individual reads in Nanopore and PacBio, respectively, supplementary table S10, Supplementary Material online). The longest repeat arrays we recovered were for (AATGG)n and 32-mers (fig. 4), some of which were over 59 kb (supplementary table S11 and fig. S12, Supplementary Material online). Last, we focused on the arrays with a single dominant motif and, depending on the species, found that at least 10–25% of all arrays were composed of a mix of different repeated motifs (supplementary table S12, Supplementary Material online). This is likely an underestimation, as we only detected overlaps in repeat annotations among the 39 most abundant repeated motifs. With PacBio, the longest repeat arrays we recovered were over 17 kb (supplementary table S11 and fig. S13, Supplementary Material online). Taken together, our results suggest frequent interspersion of perfect repeats with highly similar repeated motifs.
Fig. 4.
Box plots of lengths of (A) reads, (B) repeated motif (AATGG)n, and (C) one 32-mer recovered, from Nanopore data.
As a control of a repeat expected to be present in our data, we studied the repeat density of the telomeric (TTAGGG)n satellite using these long-read data, even though it is not 1 of the 39 most abundant repeats. The repeat density of this satellite was rather low for both technologies (the ranges for its density across species were 0.00194–0.0330 kb/M and 0.0110–0.0974 kb/M for Nanopore and PacBio, respectively, supplementary fig. S14A and B and table S3, Supplementary Material online), consistent with our findings from Illumina reads (supplementary table S3, Supplementary Material online). Nevertheless, we still found a substantial number of long (>500 bp) arrays of (TTAGGG)n in PacBio (the longest arrays were 10.4, 3.9, 7.2, and 10.4 kb for human, chimpanzee, gorilla, and Sumatran orangutan, respectively, supplementary fig. S14D, Supplementary Material online) and also some in our smaller-scale Nanopore data (the longest arrays were 0.8 kb and 4 kb for human and Bornean orangutan, respectively, supplementary fig. S14C, Supplementary Material online). Moreover, these telomeric satellite arrays were predominantly located toward the ends of reads (supplementary fig. S14, Supplementary Material online), further implying their telomeric location.
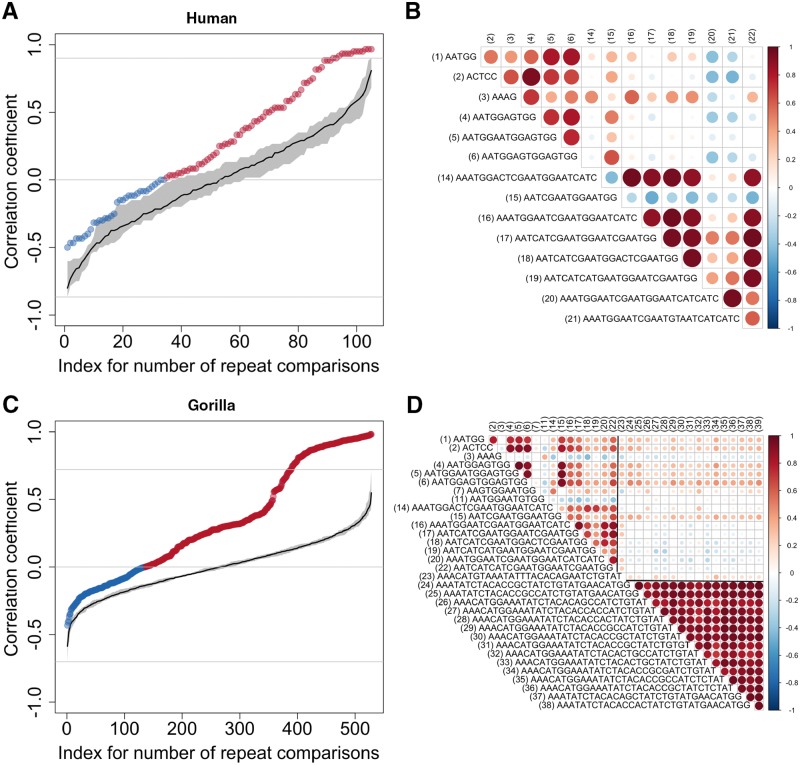
The Densities of the 39 Abundant Repeats Display High Correlations
We computed Spearman correlation coefficients for densities between pairs of repeats among the 39 abundant ones found in great ape genomes. Here, each repeat is represented by a vector of ranks for its densities across individuals (in each species). The significance of these correlations was tested against a chance background scenario simulated by random reshuffling of individuals for each repeated motif label (fig. 5 and supplementary fig. S15, Supplementary Material online; see Materials and Methods). Most correlation coefficients were positive and rather large. Furthermore, we found that blocks with strong positive correlations tended to comprise phylogenetically related repeated motifs (fig. 5 and supplementary fig. S5, Supplementary Material online). Negative and moderately large coefficients (r<−0.5) were also observed in chimpanzee, bonobo, and Sumatran orangutan (supplementary fig. S15, Supplementary Material online). In general, negative correlations were rare and mostly associated with the (AAAG)n repeat (supplementary fig. S15, Supplementary Material online).
Fig. 5.
Spearman correlations for the densities of the 39 abundant repeats in human and gorilla. Colored dots in the upper (A; Human, n = 105 comparisons) and lower (C; Gorilla, n = 528 comparisons) left panels show observed correlations between pairs of repeats plotted in nondecreasing order, in red when positive and in blue when negative. Chance background correlations, again in nondecreasing order, are plotted in black with variation bands in gray (see Materials and Methods). The heatmaps in the upper (B; Human) and lower (D; Gorilla) right panels show the correlations corresponding to each repeat pair, with various intensities of red (positive) and blue (negative). The size of the circles is also proportional to that of the correlation. Supplementary figure S9, Supplementary Material online, provides the same information for the other species. Only repeats present in the relevant species are shown.
The correlations between abundant repeat densities differed across great ape species. For example, in human, we observed many more positive correlations than expected by chance, but also a substantial number of negative correlations (fig. 5A and B). In contrast, gorilla had a sizable subset of repeats with very high and significant positive correlations (coefficients >0.8), but very few negative correlations (fig. 5C and D). In Sumatran and Bornean orangutans, we observed more positive than negative correlations—but none of the coefficients were significant (i.e., all the coefficients could have been observed by chance based on our permutation test using reshufflling of individuals for each repeated motif label; supplementary fig. S15E–H, Supplementary Material online) potentially due to small sample sizes. We found that some of the correlations between abundant repeat densities in all the species studied, in part, could be explained by motif sequence similarity and/or the physical proximity of repeated arrays (supplementary note 5, Supplementary Material online).
Discussion
Satellite repeats constitute a large portion of the human genome (Spinelli 2003; Jain, Koren, et al. 2018), yet they have been routinely underexplored in the genomes of great apes (Kronenberg et al. 2018). Our study fills this gap; it provides a detailed characterization of this important component of hominid genomes and demonstrates a remarkable divergence of satellite repeats with unit sizes ≤50 bp among ape species separated by <15 My (Glazko and Nei 2003).
Satellite Repeats in Great Ape Genomes
The (AATGG)n Repeat and Its Derivatives
We determined the (AATGG)n repeat to be abundant in great ape species. Independent of sequencing technology used, its density was usually highest in gorilla (second highest with Nanopore), rather high in orangutans, human, and bonobo, and lowest in chimpanzee (fig. 1C). This is in agreement with a suggestion that, during primate evolution, amplification of HSat3, for which the (AATGG)n repeat is the source, peaked in gorilla and orangutan lineages (Jarmuż et al. 2007). We also found high intraspecific variability in the density of (AATGG)n, sometimes reaching up to 1.51-fold pairwise difference between individuals of the same species (supplementary table S4, Supplementary Material online). These findings strongly argue for the rapid evolution of this repeat.
We found that (AATGG)n is ubiquitously present in all great ape individuals in our study, suggesting that it performs an important function. It is located at pericentromeric regions of acrocentric chromosomes (Lee et al. 1997), can fold into a non-B DNA conformation (Grady et al. 1992; Zhu et al. 1996; Chou et al. 2003), and was suggested to participate in forming centromeres (Grady et al. 1992). Importantly, under conditions of stress, the (AATGG)n repeat is transcribed from three to four 9q12 loci into long noncoding RNAs which, together with several proteins, form nuclear stress bodies and play a critical role in heat shock response (Nakahori et al. 1986; Jolly et al. 2004; Biamonti and Vourc’h 2010; Goenka et al. 2016). In fact, such RNAs were recently shown to be required to “provide full protection against the heat-shock-induced cell death” via contributing to transcriptional silencing (Goenka et al. 2016). Some of these RNAs can be very long (Jolly et al. 2004), with polyadenylated transcripts ranging from 2 to >5 kb (Goenka et al. 2016). In agreement with this observation, we found that some (AATGG)n imperfect arrays, which can be part of these transcripts, can be over 59 kb long.
Our study has also identified abundant repeated motifs that were derived from (AATGG)n (fig. 1C and supplementary fig. S5, Supplementary Material online). Interestingly, some of them, including the (AATGG)n repeat itself, are matching substrings of the most common 24-mers indicative of a specific HSat subfamily (Altemose et al. 2014)—either with no mismatches (AATGG, ACTCC, and AAAG) or with one mismatch (AATGGAATGGAGTGG, AATGGAGTGG, AATGGAATGTG, AATCGAATGGAATGG). This provides an independent confirmation that they form satellite repeats.
Subterminal Satellites
Another interesting group of satellite repeats highlighted by our study are the phylogenetically related, AT-rich 32-mers also called StSats due to their proximity to telomeres, as demonstrated by our and other studies (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012). Independent of the sequencing technology used, we found that these repeats are highly abundant in gorilla, still very abundant in chimpanzee and bonobo, but absent in human. These findings corroborate early studies hypothesizing that these repeats were present in the common ancestor of hominids (albeit in small amounts), and then lost in the human lineage (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012). The loss of StSats in orangutans was also proposed (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012), however, our analysis suggests that such loss was incomplete, as we can still find StSat traces in orangutan genomes using both Illumina and Nanopore read data. Consistent with the notion of a partial loss in orangutans, StSats are polymorphic in their presence/absence among orangutan individuals (supplementary fig. S7H, Supplementary Material online). In contrast, the majority of StSats are present in all gorilla, chimpanzee, and bonobo individuals analyzed, suggesting that they might be functionally important in their genomes. Various roles for StSats have been proposed, including participation in meiosis (Royle et al. 1994; Koga et al. 2011; Ventura et al. 2012), telomere clustering and metabolism, as well as the regulation of replication timing in the vicinity of telomeres (Novo et al. 2013).
Male-Biased Repeats
Leveraging differences in repeat density between males and females, we identified 18 candidate male-biased repeats in great apes (supplementary table S6, Supplementary Material online). These included the (AATGG)n repeat, which was previously shown to be present on the human Y chromosome as the primary repeated unit of its three common satellites (DYZ1, DYZ17, and DYZ18) (Kunkel et al. 1976; Skaletsky et al. 2003), and on the Y chromosome of orangutan, gorilla, and chimpanzee/bonobo with FISH (Jarmuż et al. 2007). Additionally, we found several StSats to be male-biased and confirmed their presence in the gorilla Y assembly and in the bonobo Y chromosome using FISH (fig. 3). This substantially increases the current knowledge of both candidate and validated Y chromosome heterochromatic repeats in great apes. Prior to our study, these repeats were underexplored because only human, chimpanzee, and gorilla Y chromosome assemblies are currently available and such assemblies are mostly euchromatic (Skaletsky et al. 2003; Hughes et al. 2010; Tomaszkiewicz et al. 2016).
It was proposed that enrichment of different, or accumulation of unique, satellite DNA is the first step in separation of the X and Y chromosomes (Brutlag 1980). It was also hypothesized that the composition of the heterochromatin on the Y may differ from that on other chromosomes because of 1) absence of recombination; 2) a potential role of heterochromatin in silencing the Y; and 3) the small effective population size of the Y (Nei 1970; Charlesworth and Charlesworth 2000; Bachtrog 2013). Consistent with these hypotheses, some Drosophila species (D. virilis, D. melanogaster, D. simulans, and D. sechellia) exhibited many Y-enriched or Y-specific satellite repeats (Wei et al. 2018). In contrast, other Drosophila species (D. pseudoobscura and D. persimilis) have prominent abundance of transposable elements (TE) on the Y (Wei et al. 2018)—suggesting that Y chromosome degeneration occurs by satellite repeat accumulation in some species, and TE accumulation in others. These two alternatives can be explored also for the great ape Y chromosomes (supplementary note 6, Supplementary Material online), once the assemblies that are currently missing become available.
The Y chromosome heterochromatin is a major source of epigenetic regulation, modulating phenotypic variation in natural populations (Lemos et al. 2010). For instance, in Drosophila, its content and length affect expression of autosomal genes (Lemos et al. 2008). Similarly, a repeat-rich noncoding RNA was recently found to play a role in regulating the expression of several genes in mouse testis (Reddy et al. 2018). Such a phenomenon in primates is yet to be investigated.
Co-Occurrence of Satellite Repeats
Our observations suggest dependencies among the densities of many repeated motifs, and an underlying structure in their distribution in the great apes genomes—which is at least partially dictated by sequence similarity and evolution, stemming from the interspersion of longer satellite arrays with similar motifs. This echoes recent observations made for Drosophila (Wei et al. 2014, 2018) and Chlamydomonas reinhardtii (Flynn et al. 2018). Similarly to the pattern observed in Drosophila, in great apes clusters of co-occurring repeats are in part driven by their sequence similarity. Several hypotheses were proposed to explain such a pattern; for instance, many similar repeat motifs can serve as recognition sites for the same DNA-binding proteins (Wei et al. 2014), and correlated motifs might be physically linked to each other due to a large-scale duplication or due to interspersion (supplementary note 5, Supplementary Material online). An example of interspersion are two groups of HSat3 DNA: the first group is dominated by (AATGG)n and the second group represents a mix of (AATGG)n and (ACTCC)n (Jarmuż et al. 2007). We also found antagonistic relationships among some repeats, in particular among (AAAG)n and several other repeats. Again similar to observations made in Drosophila (Wei et al. 2014), this can occur when the expansion of one repeat type comes at the expense of another. The differences we found in nature and strength of dependencies among repeat densities in various great apes might be explained by differences in the overall tolerance their genomes have toward repetitive load. Future studies should incorporate data on long-distance genome interactions (e.g., Hi-C) to further explore repeat co-occurrence patterns in great ape genomes.
Interspecific Differences and Lack of Phylogenetic Signal in Repeat Densities
We found drastic differences among great ape species in overall repeat content. Independent of the sequencing technology used, overall repeat density was highest in gorilla, intermediate in chimpanzee and bonobo, and lowest in human and orangutans (fig. 1C). This is primarily explained by the absence or paucity of StSats in human and orangutans, respectively. In addition, while clustering based on repeat densities did correctly assign individuals into species, subsequent agglomeration did not follow the expected species phylogeny. In particular, we frequently observed chimpanzee, bonobo, and gorilla clustering together, and human clustering with orangutan (fig. 2B). Several explanations are possible for this unexpected observation, including incomplete lineage sorting (Kronenberg et al. 2018), parallel gains of the same repeats along different lineages, molecular drive, and segregation distortion (reviewed in Wei et al. 2018). Future studies should examine each of these explanations in detail. At present, what is clear is that satellite repeats have a notably high tempo of turnover and, at least at the timescale resolution of great ape evolution, do not carry phylogenetic signals. This is in contrast with the recent observation in Drosophila, where simple satellites recapitulated the phylogenetic tree at the species level, but not at the level of populations (Wei et al. 2014, 2018).
The Power of Long Reads, Study Caveats, and Future Directions
One of the strengths of our study is in that we combined information from three different sequencing technologies to investigate satellite repeats. Importantly, these three technologies have distinct error profiles, accurate Illumina sequencing (<0.1% error rate) has at least twice more substitutions than indels, while Nanopore and PacBio are dominated by deletions and insertions, respectively (supplementary table S13, Supplementary Material online). The longest repeat arrays we identified using the Nanopore and PacBio platforms were 59 kb and 17 kb in length. Such lengths are unprecedented; the recent PacBio-augmented assembly of the sooty mangabey (a primate) identified a 52 kb repeat array, and this was the longest found in an analysis comprising as many as 719 assembled eukaryotic genomes (Surabhi et al. 2018). Our study confirms that long-read technologies are indeed suitable for the analysis of long heterochromatic satellites. This is due both to their progressively increasing read lengths, and to recent advances in the algorithms used to tackle their noisy error profiles, for example, NCRF (Harris et al. 2019). Deciphering repeat lengths and structures will enable genotyping and assigning potential functions to a larger set of repeat arrays than previously possible. For example, Sonay et al. (2015) showed gene expression divergence between human and great apes to be higher for genes that encompassed tandem repeats (TRs). However, since their study required TRs to be fully encompassed within short Illumina sequencing reads, they were able to analyze only 58% of TRs present in the human reference. Nanopore sequencing was recently used to characterize the first complete human centromere on the Y chromosome (Jain, Olsen, et al. 2018) and to determine the lengths of human telomeric repeats (Jain, Koren, et al. 2018). We expect a growing interest in tools and approaches operating directly on raw, ultra-long reads (Lower et al. 2018).
Many of our conclusions are robust to the use of sequencing technology. However, we did find differences in the exact values of repeat density estimates obtained from the three technologies we considered. These differences could be due to the use of different individuals between short- and long-read technologies, but also due to the vastly different library preparation and sequencing protocols. While Illumina reads always represent short fragmented DNA, long DNA molecules used for PacBio and Nanopore sequencing could form secondary structures. We have recently shown that non-B DNA structures can affect PacBio sequencing depth and error rates (Guiblet et al. 2018). For Nanopore, fragments harboring these structures might not pass through the pores. In both cases, the representation of repeats capable of forming non-B DNA might be altered. This, for instance, might explain at least in part why the (AATGG)n repeat, known to form a non-B DNA structure (Grady et al. 1992), is underrepresented in Nanopore and PacBio versus Illumina data (fig. 1C). The telomeric repeat (TTAGGG)n is known to adopt a G-quadruplex formation and this might also affect its low density in sequencing reads. Moreover, different genome k-mers are not represented equally in Nanopore sequencing, an issue that is being mitigated by advances in the Nanopore base calling algorithms (Ip et al. 2015; Lu et al. 2016). The Illumina short-read sequencing used in the first part of our study might have its own issues. The APD and HGDP sequencing libraries we analyzed were generated with the PCR+ protocol. This might have led to an overestimation of repeat densities or difficulties with sequencing of the extremely GC-rich fragments. However, human repeat densities were very similar when estimated from PCR+ versus PCR− samples (fig. 1C and supplementary fig. S4, Supplementary Material online), and we observed each repeat motif at each locus to be affected by PCR amplification at approximately the same rate (supplementary note 2, Supplementary Material online). In Drosophila (Wei et al. 2018), omission of the PCR step improved correlation of satellite abundances between replicates. It is much more expensive to generate PCR− data on a large scale in apes than in Drosophila, especially when intraspecific variation, and thus multiple individuals, are of interest. However, such data should definitely be generated for great apes in the future. In this study, we did not perform the GC-bias correction (Benjamini and Speed 2012) that was employed in some other studies (e.g., Flynn et al. 2017; Wei et al. 2018). Available GC-correction pipelines require reference genomes and are thus unsuitable for whole-genome sequencing reads with suboptimal or missing references (e.g., for Y chromosomes in most apes). Additionally, each orangutan species has a low sample size (N = 5), however, the repeated motifs and their densities were similar suggesting that sampling has not exaggerated their potential differences.
Our study focused on relatively short repeated units (<50 bp), because we identified satellite repeats from short reads (two 50 bp repeats fit a 100 bp read). Our use of such short-motif repeats as a proxy for heterochromatin is justified based on several considerations: 1) they are part of long arrays, as identified by long-read data; 2) some of them match to 24-mers differentiating HSat families (Altemose et al. 2014); and 3) some of them have (sub)telomeric locations, as demonstrated by our FISH experiments (fig. 3). Repeats with longer units were not considered because the computational tools to identify them de novo in noisy long reads do not currently exist. Some studies focused on the analysis of the 171-bp centromeric heterochromatic arrays whose sequence in the human genome has been well characterized (Melters et al. 2013; Miga et al. 2014; Jain, Olsen, et al. 2018). Analyzing repeats with longer repeat units in great apes will be of great interest for future studies, once algorithms to reliably identify novel repeats from noisy long reads are developed.
Materials and Methods
Sequencing Data and Quality Filtering
From the ADP (Prado-Martinez et al. 2013), we focused on 399 fastq files with forward reads because they surpassed those with reverse reads in both sample size and quality (the latter was computed using FastQC v0.11.2 for all files using ten randomly selected reads per file). Ape individuals sequenced in multiple Illumina sequencing lanes/runs were kept separately for all the downstream processing and treated as technical replicates. Excluding 39 files with read lengths shorter than 52 bp resulted in 360 files (322, 32, and 6 files with read lengths 100, 101, and 151 bp, respectively). Subsequently, excluding 51 files with read counts smaller than 20,000,000 (to avoid potential sampling bias resulting from low read counts) resulted in 309 files. The files belonging to genetically close relatives to other samples (Bulera, Kowali, Suzie, and Oko) (Prado-Martinez et al. 2013) were also removed, resulting in 295 fastq files. To avoid sequence bias revealed by QC analysis (overrepresented k-mers present profusely toward read ends) and to remove potential sequencing errors, we discarded all reads that contained at least 1 bp with a Phred quality score <20 using FASTX-Toolkit (version 0.0.13, fastq_quality_filter -Q33 -v -q 20 -p 100).
Identification of Repeats
Reads retained after such filtering were converted from fastq to fasta format and repeats in them were identified with TRF (version trf409.legacylinux64, parameters MATCH = 2 MISMATCH = 7 DELTA = 7 PM = 80 PI = 10 MINSCORE = 50 MAXPERIOD = 2000 -l 6 -f -d -h -ngs) (Benson 1999). The resulting repeats were parsed using the script parseTRFngs.py (see GitHub repository) that implements collapsing of the same group of repeats (shifts and reverse complements) into a single representative. We required each repeat array to be at least 75 bp in length. Finally, we used median repeat densities across all technical replicates to compute satellite repeat densities for each individual. To verify that technical replicates from the same individual were consistent in their repeat estimates, we measured the tightness of these estimates computing intraclass correlation coefficients between technical replicates for the 100 most abundant repeats (we used the R package ICCbare). The median intraclass correlation coefficient was 0.96 (supplementary fig. S1, Supplementary Material online).
To avoid duplicates in the output, the recovered repeats were further filtered and formatted. Namely, we merged all repeats that shared the basic repeated unit and were in close vicinity (less than the minimal unit length of the two neighboring repeats) to each other. Reads containing the same repeated motif can map to either reference or reverse strand, and the annotated repeats can start with a different leading nucleotide. Thus, we report the data on occurrences of a repeated motif whose phase was chosen alphabetically, and combine the data for motifs and their reverse complement sequences. Because the same long stretches of repeats can have different beginnings (e.g., AATGG and ATGGA differ by a 1 bp shift) or can be present on different strands (e.g., AATGG and CCATT), we reformatted all repeats into the lexicographically smallest rotations. This means that for all possible rotations (1 bp shift followed by 1 bp increments of shift size up to the unit length) and both possible strands, we picked only one representative. This representative is the first repeat in alphabetical order out of all generated possibilities that we described earlier.
Calculation of Repeat Frequency and Density
We required each repeated motif to be present at ≥100 loci per 20 million reads. For repeated motifs that passed these filters, we calculated the corresponding repeat densities after normalizing for the read length and the read count after filtering. To calculate repeat density for each species, we included only those repeats that were present in at least one individual of that species. In order to display repeat densities in the heatmaps, they were first converted to kb/Mb and then rounded to two decimal places.
Correlations of Repeat Co-Occurrences
To assess the significance of observed correlations of repeat motifs (using Spearman coefficient and ranks based on the repeat density), we generated ten reshuffled data sets of the original repeat densities of 39 abundant repeats separately for each species (visualized as gray band in fig. 5). Reshuffling was done as follows: in a matrix of individuals x repeats, we kept the content of the matrix, but randomly reassigned row names for each column, so that the biological associations among repeats were broken and those occurring were due to chance.
Sequence Similarity and Interrelatedness among the 39 Most Abundant Repeated Motifs
For supplementary figure S5, Supplementary Material online, the sequence similarity was calculated using MEGA7 (Kumar et al. 2016). Only substitutions (and not insertions or deletions) were considered. The pairwise distances were calculated using the number of differences (both transitions and transversions) and treating gaps with pairwise deletion (supplementary fig. S5, Supplementary Material online). For each species, we calculated mean repeat density across all individuals. For the distances in supplementary note 5, Supplementary Material online, we aimed to calculate the smallest number of possible mutational steps between two motifs. Therefore, we aligned the shorter motif to a longer motif in a way that minimizes the possible differences (alignment was performed using R package msa, see script sequence_similarity_versus_cooccurrence.Rnw). We also considered all possible rotations and the reverse complement of the shorter motifs and used the global minimum.
Length Distribution for Long Reads
Repeated motifs were identified in long reads using NoiseCancelingRepeatFinder, version 0.09.03 (Harris et al. 2019). The current version of the algorithm can be downloaded from: https://github.com/makovalab-psu/NoiseCancellingRepeatFinder. For PacBio and Nanopore sequencing, –scoring=pacbio (M = 10 MM = 35 IO = 33 IX = 21 DO = 6 DX = 28) and –scoring=nanopore (M = 10 MM = 63 IO = 51 IX = 98 DO = 27 DX = 34) options were used, respectively. The maxnoise parameter was set to 20% to retain long reads with noisy repeat arrays. Subsequently, the repeated arrays were analyzed for their motif composition and each array was assigned to a motif that comprises >50% of an array. The following settings were used to run NCRF: –scoring=nanopore/pacbio –stats=events –positionalevents –maxnoise=20% –minlength=75; Filtering step: ncrf_consensus_filter.py –winner=0.5.
Experimental Validations of Male-Biased Repeats
Preparation of the Probes
The WBY painting probe was prepared from flow-sorted bonobo Y chromosomes and labeled with biotin-16-dUTP (Jena BioScience) using DOP-PCR according to (Yang et al. 2009). Oligonucleotide probe (Pan32) (/5AmMC12/ATCTGTATAAACATGGAAATATCTACACCGCY) was prepared and labeled using Alexa Fluor oligonucleotide amine labeling kit (Invitrogen).
Fluorescent In Situ Hybridization
Metaphases were prepared from chimpanzee male lymphoblastoid cell line and from bonobo male fibroblast cell line following a standard protocol of colcemid treatment, hypotonization and methanol/acetic acid fixation (Howe et al. 2014). Slides were pretreated with acetone for 10 min and aged at 65 °C for 1 h. Subsequently, the slides were denatured in the alkaline solution (Sigma) for 5 min, followed by neutralization in 1 M Tris–HCl, pH 7.5, and one wash in 1× PBS for 4 min. Next, a series of dehydration washes were performed as follows: 70% EtOH at −20 °C for 4 min, 70% EtOH for 2 min, 90% EtOH for 2 min, and 100% EtOH for 4 min. The WBY probe was denatured in hybridization buffer at 75 °C for 15 min and preannealed at 37 °C for 30 min. Subsequently, 25 ng of the Pan32 probe was applied to the hybridization area and incubated at 37 °C for 12 h for chimpanzee male chromosomes as well as for bonobo male chromosomes. In a separate FISH experiment, the mix of 25 ng of WBY and 25 ng of Pan32 was applied to the hybridization area and incubated at 37 °C for 24 h for bonobo male chromosomes and for 48 h for chimpanzee male chromosomes (cross-species FISH). Posthybridization washes were performed in 0.5× SSC at 50 °C for 5 min, 2× SSCT at 37 °C for 5 min, and 1× PBS at 37 °C for 5 min. For slides with the mix of probes, an additional step of probe detection with Cy3-Streptavidin (Sigma) was applied. Slides were stained with DAPI (Vector Laboratories) and visualized under the Keyence BZ-9000 fluorescence microscope. Photodocumentation was performed using the 100× immersion objective and the images were analyzed using BZ-Viewer and BZ Analyzer.
Nanopore Library Preparation and Sequencing
DNA was extracted from male cell lines of bonobo (AG05253, Coriell Institute), gorilla (KB3781, “Jim,” San Diego Zoological Society), Bornean orangutan (AG05252, Coriell Institute), and Sumatran orangutan (AG06213, Coriell Institute) using the MagAttract High Molecular Weight DNA Kit (Qiagen, Germany). Male chimpanzee DNA sample (CH159, “Rock”) was provided by Dr. Mark Shriver and was acquired from the Bastrop Research Center. Human male DNA (J101) was provided by the University of Chicago.
Residual RNA was removed by digesting 3.5 µg of extracted DNA with 10 µg RNase A (Amresco) at 37 °C for 1 h, followed by purification with 1 volume of AMPure XP beads (Beckman Coulter). DNA integrity was visualized on a 0.5% agarose gel, DNA purity was determined with NanoDrop, and the concentration was measured with a Qubit broad-range assay. Libraries were prepared with the Native Barcoding Kit 1 D (PCR-free) and the Ligation Sequencing Kit 1 D (Nanopore) starting with 2 µg DNA per sample. DNA repair and end-repair were combined in one step as described in the 1 D gDNA long reads without BluePippin protocol (version: GLRE_9052_v108_revB_19Dec2017; updated: January 10, 2018). Barcoding and adapter ligation were performed as indicated in the 1 D Native barcoding genomic DNA (with EXP-NBD103 and SQK-LSK108) protocol (version: NBE_9006_v103_revP_21Dec2016; updated: February 16, 2018), starting with 700 ng of end-prepped DNA per sample. About 250 ng of barcoded DNA per sample were pooled and all further steps were performed according to the 1 D gDNA long reads without BluePippin protocol. DNA low binding tubes as well as wide-pore low-retention pipette tips were used for DNA handling in all steps. Sequencing was performed with a MinION using a flow cell of the type FLO-MIN106—R9.4 for 48 h. This resulted in 396, 55, 667, 526, 615, and 383 Mb of data (distributed among 26, 4, 43, 36, 40, and 22 thousand reads) for human, chimpanzee, bonobo, gorilla, Sumatran orangutan, and Bornean orangutan, respectively.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Marzia Cremona, Kate Anthony, Oliver Ryder, Mark Shriver, Malcolm Ferguson-Smith, Jorge Pereira, and Shaun Mahony for their assistance. We thank Wilfried Guiblet and Arslan Zaidi for valuable biological insights. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM130691. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding was also provided by the Eberly College of Sciences, The Huck Institute of Life Sciences, and the Institute for CyberScience, at Penn State, as well as, in part, under grants from the Pennsylvania Department of Health using Tobacco Settlement and CURE Funds. The department specifically disclaims any responsibility for any analyses, responsibility, or conclusions.
References
- 1000 Genomes Project Consortium Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 5267571:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Hasson D, Cheung F, Warburton PE.. 2010. A paucity of heterochromatin at functional human neocentromeres. Epigenet Chromatin. 31:6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altemose N, Miga KH, Maggioni M, Willard HF.. 2014. Genomic characterization of large heterochromatic gaps in the human genome assembly. PLoS Comput Biol. 105:e1003628.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 142:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, Cande WZ.. 2000. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 113(Pt 6):1033–1042. [DOI] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, Zaret KS.. 2016. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 321:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Speed TP.. 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 4010:e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 272:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Vourc’h C.. 2010. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 26:a000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachary M, Guilmatre A, Quilez J, Hasson D, Borel C, Warburton P, Sharp AJ.. 2014. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet. 106:e1004418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag DL. 1980. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 14:121–144. [DOI] [PubMed] [Google Scholar]
- Calderón M, del C, Rey M-D, Cabrera A, Prieto P.. 2014. The subtelomeric region is important for chromosome recognition and pairing during meiosis. Sci Rep. 4:6488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. 2002. A human genome diversity cell line panel. Science 2965566:261–262. [DOI] [PubMed] [Google Scholar]
- Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M.. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 5177536:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 2000. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 3551403:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SH, Chin KH, Wang AH 2003. Unusual DNA Duplex and Hairpin Motifs. Nucleic Acids Res. 31(10):2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 710:e1000234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Caldas I, Cristescu ME, Clark AG.. 2017. Selection constrains high rates of tandem repetitive DNA mutation in Daphnia pulex. Genetics 2072:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Lower SE, Barbash DA, Clark AG.. 2018. Rates and patterns of mutation in tandem repetitive DNA in six independent lineages of Chlamydomonas reinhardtii. Genome Biol Evol. 107:1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Cohen EH, Polan ML.. 1971. Repetitive DNA sequences in Drosophila. Chromosoma 333:319–344. [DOI] [PubMed] [Google Scholar]
- Gläser B, Grützner F, Willmann U, Stanyon R, Arnold N, Taylor K, Rietschel W, Zeitler S, Toder R, Schempp W.. 1998. Simian Y chromosomes: species-specific rearrangements of DAZ, RBM, and TSPY versus contiguity of PAR and SRY. Mamm Genome. 93:226–231. [DOI] [PubMed] [Google Scholar]
- Glazko GV, Nei M.. 2003. Estimation of divergence times for major lineages of primate species. Mol Biol Evol. 203:424–434. [DOI] [PubMed] [Google Scholar]
- Goenka A, Sengupta S, Pandey R, Parihar R, Mohanta GC, Mukerji M, Ganesh S.. 2016. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J Cell Sci. 12919:3541–3552. [DOI] [PubMed] [Google Scholar]
- Goodman M, Grossman LI, Wildman DE.. 2005. Moving primate genomics beyond the chimpanzee genome. Trends Genet. 219:511–517. [DOI] [PubMed] [Google Scholar]
- Gordon D, Huddleston J, Chaisson MJP, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW.. 2016. Long-read sequence assembly of the gorilla genome. Science 3526281:aae0344.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen JW, Gay EH.. 1933. Effect of temperature on eversporting eye color in Drosophila melanogaster. Science 771995:312.. [DOI] [PubMed] [Google Scholar]
- Grady DL, Ratliff RL, Robinson DL, McCanlies EC, Meyne J, Moyzis RK.. 1992. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 895:1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiblet W, Cremona M, Cechova M, Harris R, Kejnovska I, Kejnovsky E, Eckert KA, Chiaromonte F, Makova KD.. 2018. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 2812:1767.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Cechova M, Makova K.. 2019. Noise-cancelling Repeat Finder: uncovering tandem repeats in error-prone long-read sequencing data. Bioinformatics (accepted on June 5, 2019). [DOI] [PMC free article] [PubMed]
- Hayden KE, Strome ED, Merrett SL, Lee H-R, Rudd MK, Willard HF.. 2013. Sequences associated with centromere competency in the human genome. Mol Cell Biol. 334:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe B, Umrigar A, Tsien F.. 2014. Chromosome preparation from cultured cells. J Vis Exp. e50203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SKM, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 4637280:536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip CLC, Loose M, Tyson JR, de Cesare M, Brown BL, Jain M, Leggett RM, Eccles DA, Zalunin V, Urban JM, et al. 2015. MinION Analysis and Reference Consortium: phase 1 data release and analysis. F1000Res. 4:1075.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT, et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 364:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH.. 2018. Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol. 364:321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuż M, Glotzbach CD, Bailey KA, Bandyopadhyay R, Shaffer LG.. 2007. The evolution of satellite III DNA subfamilies among primates. Am J Hum Genet. 803:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C.. 2004. Stress-induced transcription of satellite III repeats. J Cell Biol. 1641:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kit S. 1961. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 3:711–716. [DOI] [PubMed] [Google Scholar]
- Koga A, Notohara M, Hirai H.. 2011. Evolution of subterminal satellite (StSat) repeats in hominids. Genetica 1392:167–175. [DOI] [PubMed] [Google Scholar]
- Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, Underwood JG, Nelson BJ, Chaisson MJP, Dougherty ML.. 2018. High-resolution comparative analysis of great ape genomes. Science 360(6393) Available from: 10.1126/science.aar6343, last accessed July 05, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel LM, Smith KD, Boyer SH.. 1976. Human Y-chromosome-specific reiterated DNA. Science 1914232:1189–1190. [DOI] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, et al. 2000. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 2885466:665–669. [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC 1997. Human Centromeric DNAs. Human Genetics. https://doi.org/10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Hartl DL.. 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 3195859:91–93. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL.. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 10736:15826–15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Brutlag DL.. 1987. Identical satellite DNA sequences in sibling species of Drosophila. J Mol Biol. 1942:161–170. [DOI] [PubMed] [Google Scholar]
- Lower SS, McGurk MP, Clark AG, Barbash DA.. 2018. Satellite DNA evolution: old ideas, new approaches. Curr Opin Genet Dev. 49:70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Giordano F, Ning Z.. 2016. Oxford nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinformatics. 145:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. 1978. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma 661:23–32. [DOI] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 141:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 3386104:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ.. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 244:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahori Y, Mitani K, Yamada M, Nakagome Y.. 1986. A human Y-chromosome specific repeated DNA family (DYZ1) consists of a tandem array of pentanucleotides. Nucleic Acids Res. 1419:7569–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1970. Accumulation of nonfunctional genes on sheltered chromosomes. Am Nat. 104938:311–322. [Google Scholar]
- Novo C, Arnoult N, Bordes W-Y, Castro-Vega L, Gibaud A, Dutrillaux B, Bacchetti S, Londoño-Vallejo A.. 2013. The heterochromatic chromosome caps in great apes impact telomere metabolism. Nucleic Acids Res. 419:4792–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409(1–2):72–82. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 4997459:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, Joshi RS, Mittelman D, Sharp AJ.. 2016. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res. 448:3750–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy HM, Bhattacharya R, Jehan Z, Mishra K.. 2018. Y chromosomal noncoding RNA regulates autosomal gene expression via piRNAs in mouse testis. bioRxiv [Internet]. Available from: https://www.biorxiv.org/content/early/2018/03/24/285429.abstract, last accessed July 05, 2019. [DOI] [PMC free article] [PubMed]
- Rhoads A, Au KF.. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 135:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S, Raza ST, Mahdi F.. 2014. Telomere length variations in aging and age-related diseases. Curr Aging Sci. 73:161–167. [DOI] [PubMed] [Google Scholar]
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G.. 2004. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 152:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW.. 2002. Genetic structure of human populations. Science 2985602:2381–2385. [DOI] [PubMed] [Google Scholar]
- Rošić S, Köhler F, Erhardt S.. 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol. 2073:335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle NJ, Baird DM, Jeffreys AJ.. 1994. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat Genet. 61:52–56. [DOI] [PubMed] [Google Scholar]
- Seong K-H, Li D, Shimizu H, Nakamura R, Ishii S.. 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 1457:1049–1061. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 4236942:825–837. [DOI] [PubMed] [Google Scholar]
- Sonay TB, Carvalho T, Robinson M.. 2015. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 25(11):1591–1599. [DOI] [PMC free article] [PubMed]
- Soufi A, Donahue G, Zaret KS.. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 1515:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli G. 2003. Heterochromatin and complexity: a theoretical approach. Nonlinear Dynamics Psychol Life Sci. 74:329–361. [DOI] [PubMed] [Google Scholar]
- Stephens ZD, Iyer RK.. 2018. Measuring the mappability spectrum of reference genome assemblies In: Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. BCB ’18. New York (NY): ACM; p. 47–52. [Google Scholar]
- Subramanian S, Mishra RK, Singh L.. 2003. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 42:R13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. 1961. Variation and heterogeneity of base composition of deoxyribonucleic acids: a compilation of old and new data. J Mol Biol. 31:31–IN15. [Google Scholar]
- Sujiwattanarat P, Thapana W, Srikulnath K, Hirai Y, Hirai H, Koga A.. 2015. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci Rep. 510315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surabhi S, Avvaru AK, Sowpati DT, Mishra RK. 2019. Patterns of Microsatellite Distribution across Eukaryotic Genomes. BMC Genomics 20(1):153. [DOI] [PMC free article] [PubMed]
- Tagarro I, Fernández-Peralta AM, González-Aguilera JJ.. 1994. Chromosomal localization of human satellites 2 and 3 by a FISH method using oligonucleotides as probes. Hum Genet. 934:383–388. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S.. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 211:36–44. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I.. 2010. Stress-induced activation of heterochromatic transcription. PLoS Genet. 610:e1001175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszkiewicz M, Rangavittal S, Cechova M, Sanchez RC, Fescemyer HW, Harris R, Ye D, O’Brien PCM, Chikhi R, Ryder OA, et al. 2016. A time- and cost-effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 264:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Sajjadian S, Vives L, Sudmant PH, Marques-Bonet T, Graves TA, Wilson RK, Eichler EE.. 2012. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 226:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PM. 1971. Origin of satellite DNA. Nature 2295283:306–308. [DOI] [PubMed] [Google Scholar]
- Wei K-C, Grenier JK, Barbash DA, Clark AG.. 2014. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc Natl Acad Sci U S A. 11152:18793–18798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K-C, Lower SE, Caldas IV, Sless TJS, Barbash DA, Clark AG.. 2018. Variable rates of simple satellite gains across the drosophila phylogeny. Mol Biol Evol. 354:925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Trifonov V, Ng BL, Kosyakova N, Carter NP.. 2009. Fluorescence in situ hybridization (FISH)—application guide. In: Liehr T, editor. Generation of paint probes by flow-sorted and microdissected chromosomes.Heidelberg (Berlin: ): Springer Berlin Heidelberg; p. 35–52. [Google Scholar]
- Yunis JJ, Yasmineh WG.. 1971. Heterochromatin, satellite DNA, and cell function. Science 1744015:1200–1209. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al. 2015. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 3486239:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason C, Alexander N, et al. 2016. Extensive Sequencing of Seven Human Genomes to Characterize Benchmark Reference Materials. Scientific Data 3 (June): 160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chou SH, Reid BR 1996. A Single G-to-C Change Causes Human Centromere TGGAA Repeats to Fold Back into Hairpins. Proc Natl Acad Sci U S A 93(22):12159–12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.