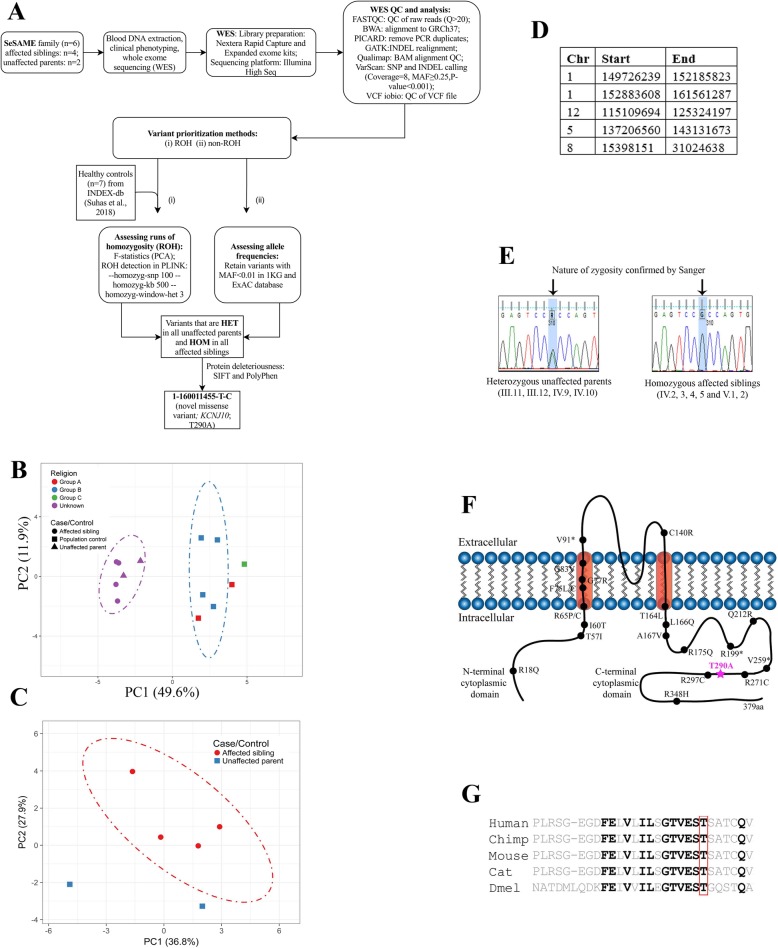
Fig. 2.
Identification of a novel mutation in KCNJ10 by homozygosity mapping and whole exome analysis of SeSAME family members. a WES analysis pipeline and variant prioritization methods. b Principle component analysis (PCA) of exome-wide F-statistics explains for an overall variance of ~ 49% (PC1) between the SeSAME family members (purple ellipse) and healthy population controls (blue ellipse). The dot-dash lines in the plot represent the 95% confidence ellipse. c PCA plot explaining intra-familial levels of homozygosity between affected and unaffected members. d ROH regions observed in all patients but not in parental controls. e The zygosity of the KCNJ10T290A variant was validated in all the six affected (HOM) and the four unaffected individuals (HET) within the pedigree. f A schematic reconstruction of Kir4.1with the T290A variant (purple) mapped in the cytoplasmic C-terminal domain, along with other deleterious variants identified from previous studies. g Multiple sequence alignment (MSA) of the Kir4.1 protein sequence across species reveals the evolutionary conservation of T290A in VEST domain