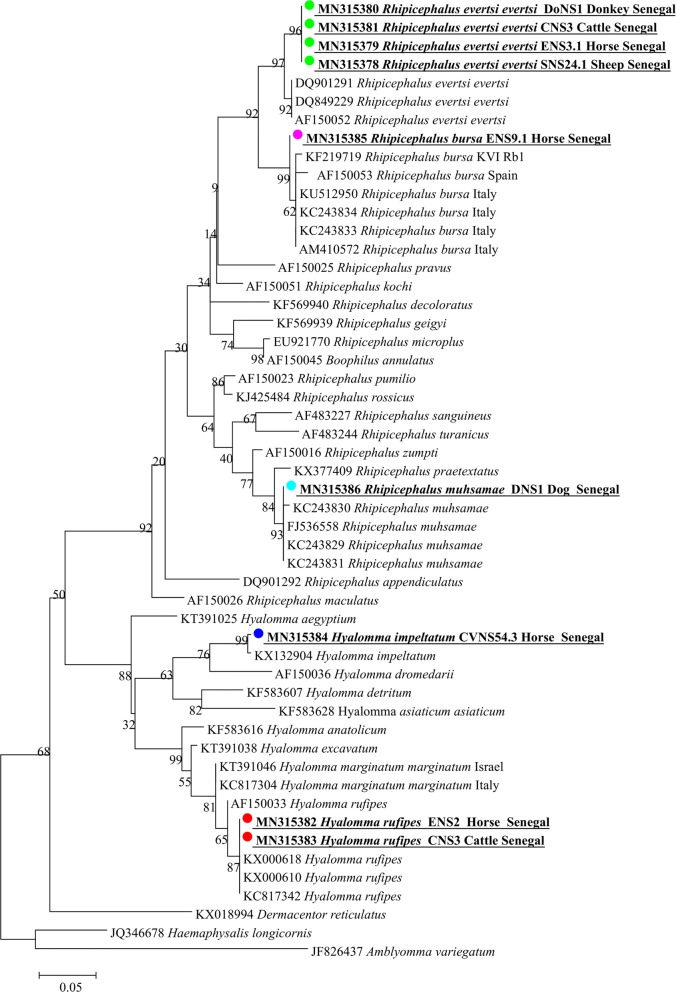
Fig. 2.
Phylogenetic tree showing the position of R. evertsi evertsi, Hy. rufipes, Hy. impeltatum, R. bursa and R. muhsamae compared to other tick species. Evolutionary analyses were conducted using MEGA7 [26]. The sequences of the 12S rDNA amplified in this study with other 12S rDNA tick sequences available on GenBank were aligned using CLUSTAL W implemented on BioEdit v.3 [25] (there were 262 positions in the final dataset). The evolutionary history was inferred by using the maximum likelihood method based on the Hasegawa–Kishino–Yano model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations. A discrete gamma distribution was used to model evolutionary rate differences among sites [2 categories (+G, parameter = 0.4726)]. The analysis involved 52 nucleotide sequences. All positions containing gaps and missing data were excluded. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The scale-bar represents a 5% nucleotide sequence divergence