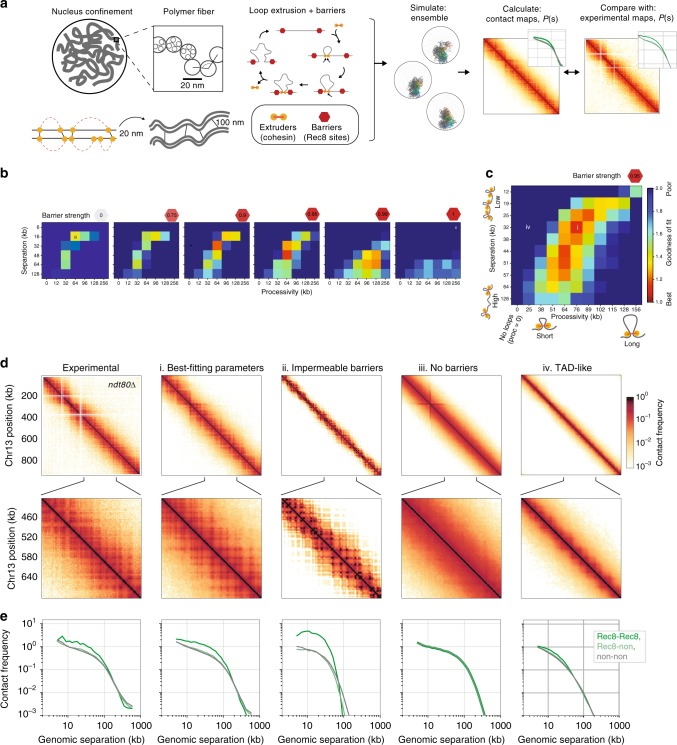
Fig. 3.
Modelling meiotic chromosome compaction. a In simulations, yeast Chr13 was represented as a polymer fibre confined to the nucleus subject to additional meiosis-specific constraints. These include extruded loops, sister crosslinks and homologue crosslinks (Methods). Barriers to extruded loops were placed at Rec8 sites30. We imposed inter-sister and inter-homologue crosslinks at sites of extruded loop bases in order to approximate the paired arrangement of homologues at pachytene. For each set of extruded loop parameters (processivity, separation and barrier strength), conformations were collected and used to generate simulated contact maps. Roughly, processivity dictates the size of an extruded loop unimpeded by collisions, separation controls the number of active extruders on the chromosome, and barrier strength controls the probability that an extruder gets paused when attempting to step past a barrier. Goodness-of-fit was then evaluated using the combined average fold discrepancy between P(s) curves for Rec8-Rec8, Rec8-non and non-non bin pairs at 2 kb resolution. Note that a value of 1 indicates perfect agreement between simulations and experimental data. b Goodness-of-fit for indicated barrier strengths over coarse grids of processivity and separation demonstrate that intermediate barrier strengths are required to agree with experimental ndt80∆ Hi-C maps. c Goodness-of-fit for a fine grid of processivity versus separation at barrier strength 0.95 (and for 0.90; Supplementary Fig. 5d). Best-fitting models had separation ~32 kb and processivity ~76 kb, corresponding to ~64% coverage of the genome by extruded loops of average length 26 kb. d. From left to right: contact maps for chromosome 13 for experimental data, and simulations with (i) best-fitting parameters, (ii) relatively stable loops between neighbouring Rec8 sites, (iii) no barriers, (iv) square TAD-like patterns of enriched contacts. Positions for each of these parameter sets indicated in b, c. e P(s) split by Rec8-Rec8, Rec8-non and non-non, as in Fig. 2e