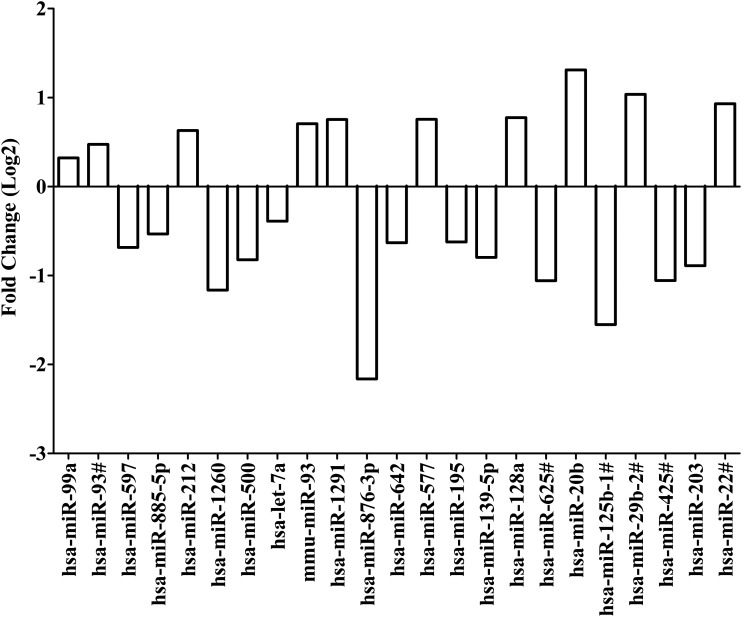
FIG. 4.
Fold change for differentially expressed microRNAs after treatment with rifampicin based on quantile normalization followed by limma differential expression analysis using the R/Bioconductor package “Automated Analysis of High-Throughput qPCR Data.” Differentiated HepaRG cells were treated with efavirenz (6.4 μM), rifampicin (24.4 μM), or DMSO (0.02%) for 24 h (including three biological replicates). Expression of microRNAs were assessed by using the TaqMan OpenArray Human MicroRNA Panel and QuantStudio 12K Flex system. The fold change in microRNA expression is compared for rifampicin versus DMSO. Quantile normalization is used to minimize variability between TaqMan OpenArray Human MicroRNA Panels and assumes that most microRNAs are not differentially expressed. Limma analysis involves fitting of a one-factorial linear model for each microRNA between rifampicin- and DMSO-treated replicates and the standard errors are moderated using an empirical Bayes model resulting in moderated t-statistics for each microRNA.