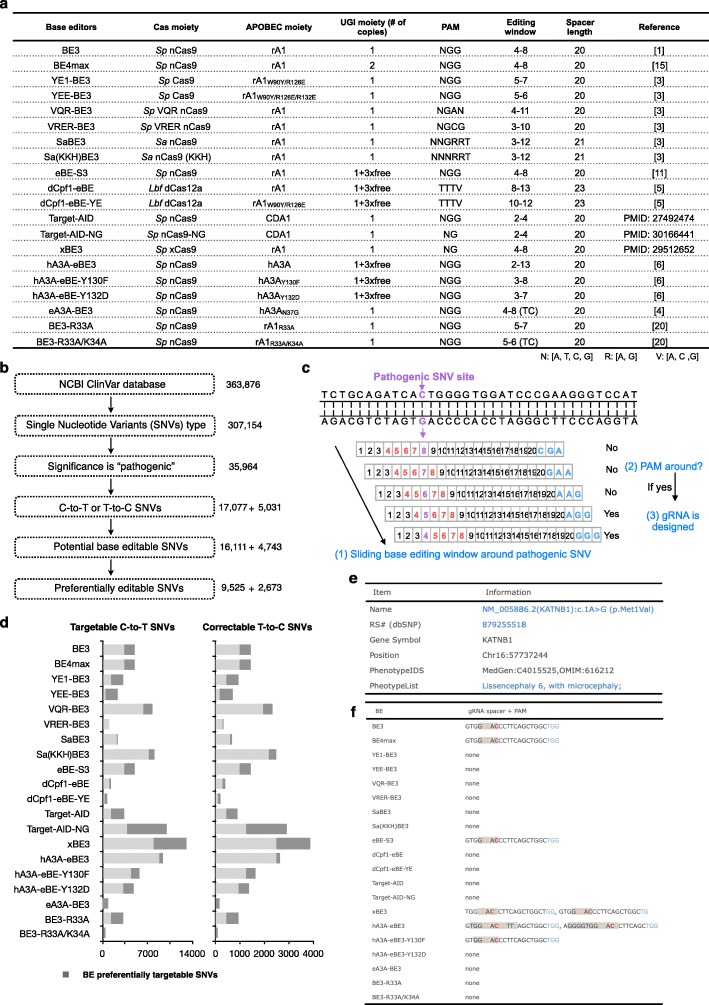
Fig. 2.
In silico base editable landscape of pathogenic SNVs. a Summary of 20 selected BEs used in BEable-GPS. PAM, editing window, spacer length, and reference are listed. b The pipeline for filtering BE editable pathogenic SNVs from NCBI ClinVar database. The numbers of variants are listed in the right for each filtering step. c The workflow of judging whether a pathogenic SNV is potentially targetable. If there are nearby PAM sequences when a SNV is in the editing window, this SNV is considered to be a potentially targetable site by BEs. d Statistics of number distribution of targetable or preferentially targetable pathogenic SNVs by each BE summarized in a. e The information of one representative pathogenic SNV is provided in the BEable-GPS online website. f The gRNA spacer region and PAM sequence of each BE for one representative SNV are shown. Pathogenic SNV in red, bystander editing site in yellow, editing window in gray, and PAM in blue