Fig. 11.
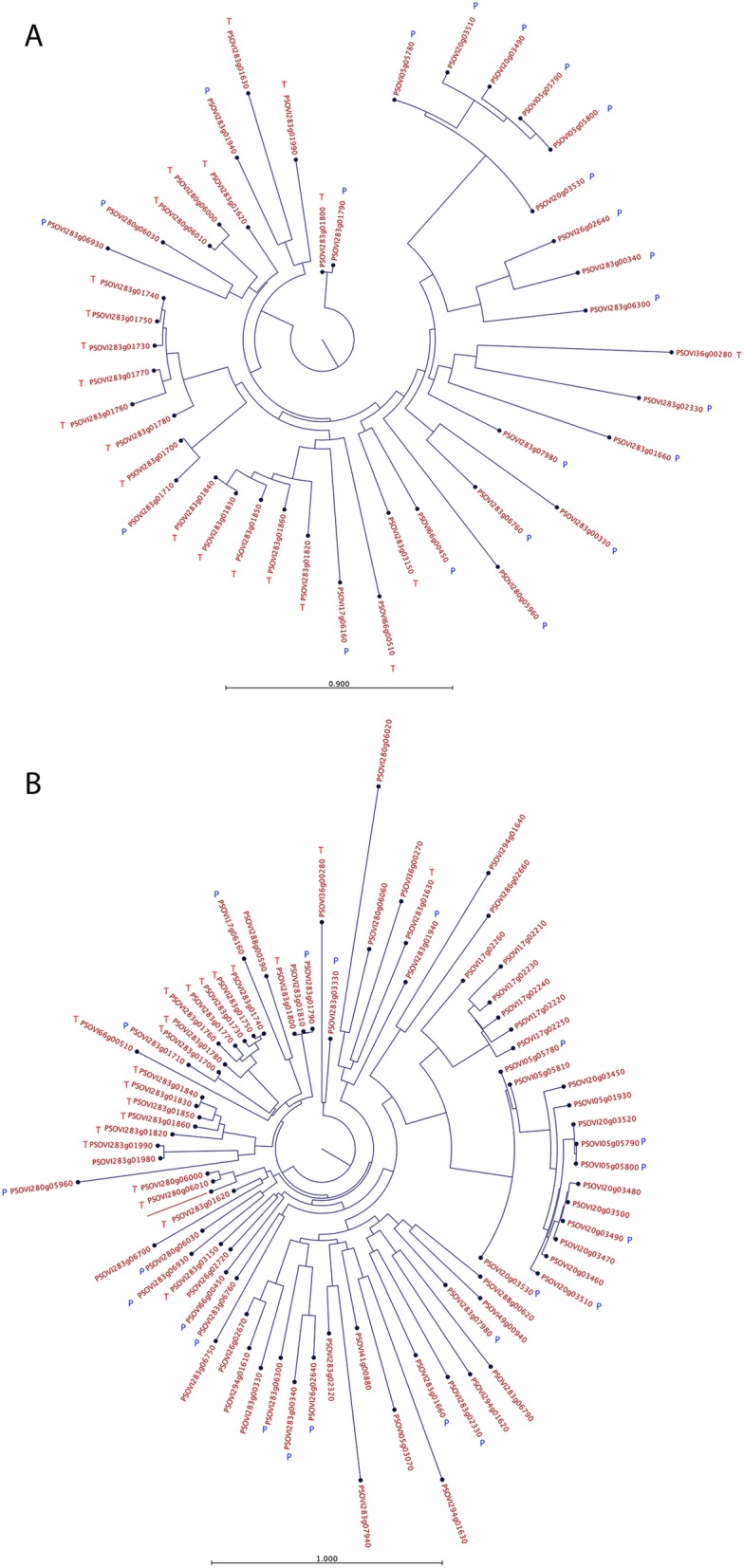
Phylogenetic analysis of DFP2-like proteins from P. ovis. Trees were constructed using a maximum likelihood phylogeny method with neighbour joining tree construction and Jukes-Cantor distance measure with 100 bootstraps. a DFP2-like genes differentially expressed (n = 44) between protonymph and tritonymph stages. b The 81 predicted P. ovis DFP2-like protein sequences from the draft P. ovis genome. DFP2-like genes upregulated in tritonymphs (vs. protonymphs) are highlighted with a red T; whilst those upregulated in protonymphs (vs. tritonymphs) are highlighted with a blue P
