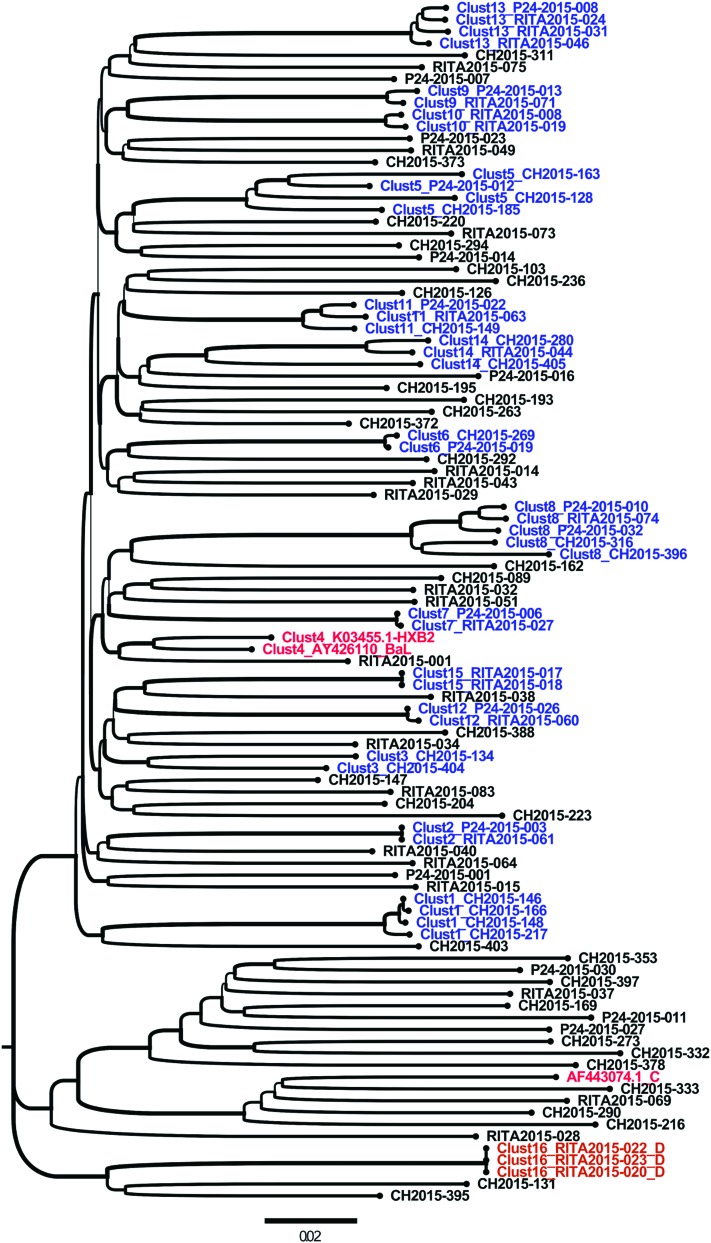
FIG. 2.
Molecular phylogenetic reconstruction of HIV-1 TCs among newly and chronically HIV-infected individuals in 2015 in Quebec using partial env-derived sequences (1,070 bp). The cluster tree contains 101 sequences from HIV-1-infected individuals, including 98 from the study cohort and 3 reference sequences introduced in the analyses as controls (AF443074.1_C, AY426110_BaL, and K03455.1-HXB2). A total of 42 HIV-1 individuals' sequences are grouped into 16 distinct small HIV-1 TCs. These clusters are labeled from Clust1 to Clust16 and are depicted using different colors. The cohort-derived clusters are shown in blue for subtype B (Clust1 to Clust15, except Clust4) and orange for subtype D (Clust16). The control sequence names are shown in red (Clust4). Sequences that did not form a cluster are shown in black. Each depicted cluster satisfied ≥99% support on bootstrap resampling of 1,000 replicates and 10% of pairwise distance.21 CH, chronic HIV-1 infection sequences; p24, recent infection status determined by p24 antigen positivity; RITA, recent infection status determined by RITA29; TCs, transmission clusters. Color images are available online.