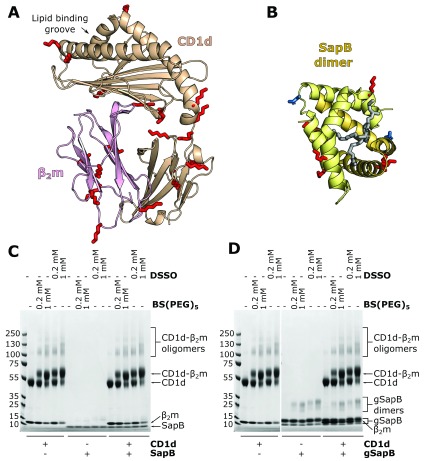
Figure 3. In vitro cross-linking of CD1d-β 2m and SapB.
A. Ribbon diagram of the structure of CD1d (beige) and β 2m (pink)(PDB ID 3U0P) with the lipid antigen groove labelled (arrow). Surface-accessible lysine residues available for cross-linking are highlighted (red sticks). B. Ribbon diagram of the structure of a SapB dimer (yellow and orange, PDB ID 1N69) with bound lipid (in gray). Lysine residues are highlighted as in panel A. The N-linked glycosylation site of SapB is highlighted (blue sticks). C. Coomassie-stained SDS-PAGE of CD1d-β 2m and unglycosylated SapB samples following incubation with cross-linkers of different lengths: DSSO (10.3 Å) or BS(PEG) 5 (21.7 Å). D. As for panel C but using CD1d-β 2m and glycosylated SapB (gSapB).