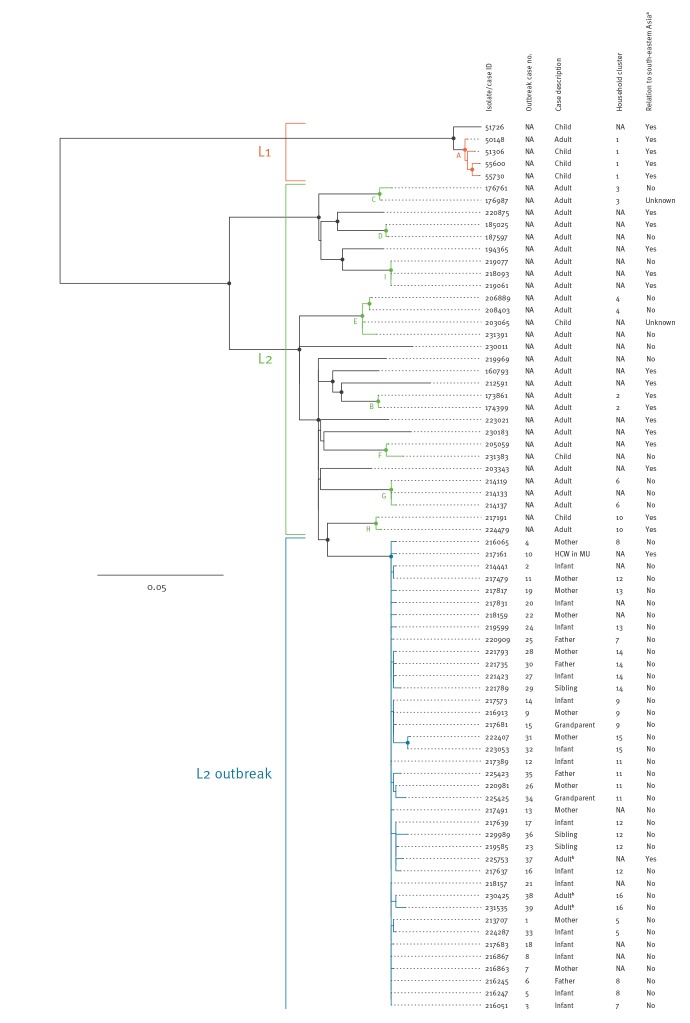
Figure 2.
Maximum-likelihood phylogeny of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus clonal complex 398 isolates, Denmark, 2006–2017 (n = 73)
HCW: healthcare worker; MU: maternity unit; NA: not applicable.
a Relation to south-eastern Asia comprises cases who were either born in or had recently visited south-eastern Asia or had known contact with such individuals.
b The three community cases identified through review of the national MRSA database.
The phylogeny was estimated from 1,071 high-quality core single nucleotide polymorphisms (SNPs) among the 73 isolates after recombination was removed from the SNP alignment, by using a general time reversible (GTR) model of nt substitution. Coloured branches indicate phylogenetic clusters (A-I) containing L1 and L2 isolates that differed by no more than 15 SNPs, which corresponds to the maximum number of pairwise SNP differences among the L2 outbreak isolates. Bootstrap values above 90% are illustrated by filled circles at the nodes. The scale bar represents the number of nt substitutions per variable site.