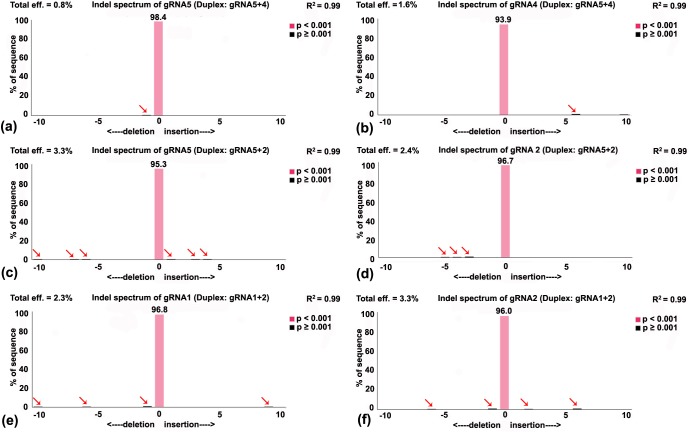
Fig 6. Indel spectrum analysis through TIDE programme.
Analyses showed probability of presence of NHEJ generated mutated sequences in upstream or downstream to the PAM motifs in ChiLCV genome after treating with three promising duplex gRNA-Cas9 constructs. Three replicates of the viral genomic sequences from individual plant were identical and hence analysis was done with one sample. Indel spectra generated surrounding the cleavage site of (a) gRNA5 (duplex: gRNA5+4), (b) gRNA4 (duplex: gRNA5+4), (c) gRNA5 (duplex: gRNA5+2), (d) gRNA2 (duplex: gRNA5+2), (e) gRNA1 (duplex: gRNA1+2), (f) gRNA2 (duplex: gRNA1+2). The analysis showed in all combinations no significant escape mutants were present. Arrow indicated insignificant occurrence of mutated sequences surrounding the PAM motif of each gRNA targeting region of ChiLCV.