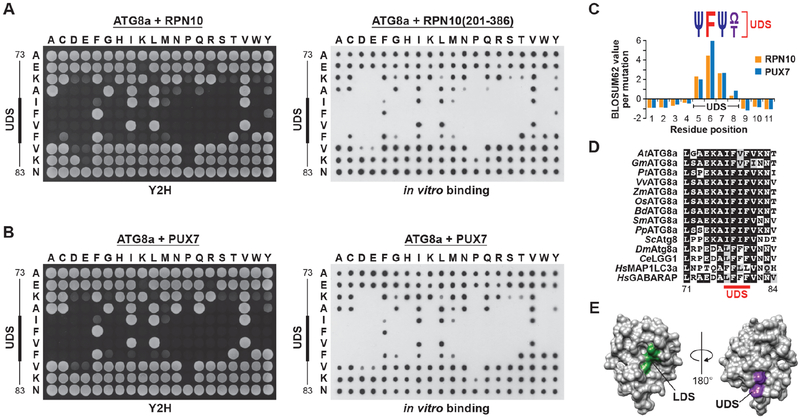
Figure 3. The UDS Contains a Conserved Phenylalanine Surrounded by Hydrophobic Residues.
(A and B) Amino acid sequence specificity of the ATG8 UDS for binding to UIMs. A panel of ATG8a mutants generated by saturating mutagenesis was tested for binding to Arabidopsis RPN10 (panel (A)) or PUX7 (panel (B)) by Y2H (left side) or dot blot binding assays (right side). Control interactions and expressions are shown in Figure S3.
(C) Quantification of the binding assays in panels (A) and (B), and comparison with expected substitution rates, identifies a core UDS sequence containing an invariant phenylalanine bracketed by hydrophobic amino acids. Ψ, small hydrophobic residues; Ω, aromatic residues.
(D) Sequence alignment of the UDS region from plant, fungi and animal ATG8 proteins. The core UDS is indicated by the solid red line.
(E) The 3-dimensional structure of yeast Atg8 (PDB file 3VXW) highlighting the opposed positions of the LDS (green) and UDS (purple).