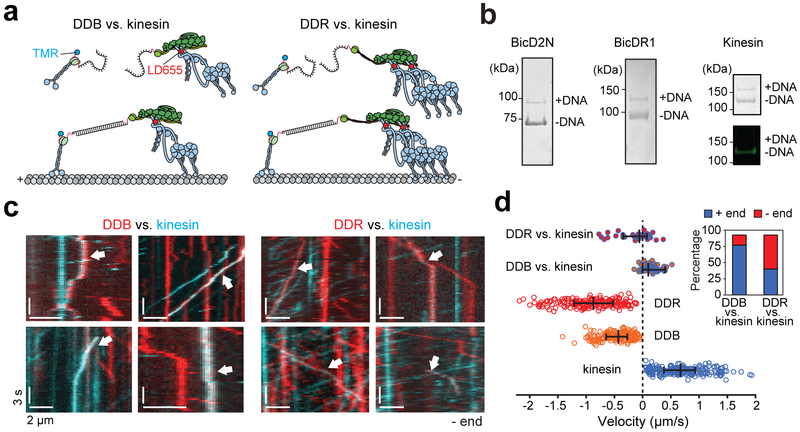
Figure 6. Recruitment of a second dynein shifts the balance towards the MT minus-end in a tug-of-war.
(a) Kinesin and BicDs were labeled with complementary DNA oligos and different-colored fluorescent dyes. Kinesin and dynein assemblies were connected by DNA hybridization. (b) (Left) The fraction of BicDs labeled with DNA was estimated from a denaturing gel. (Right) Kinesin was sub-stoichiometrically labeled with DNA (top). Fluorescence image of the same gel shows that DNA-labeled kinesin does not get labeled with TMR (bottom). n = 3 independent experiments. Full gels can be found at Supplementary Fig. 5c-d. (c) Kymographs represent the motility of TMR-kinesin (cyan), LD655-dynein (red), or kinesin-dynein assemblies (white arrows). (d) Velocity distribution of individual motors and kinesin-dynein assemblies. Negative velocities correspond to the movement towards the MT minus-end (n = 36, 46, 193, 179, and 210 from top to bottom, three independent experiments per condition). The line and whiskers represent the median and 65% c.i., respectively. The inset shows the percentage of plus- and minus-end directed runs when DDB or DDR pitted against kinesin.