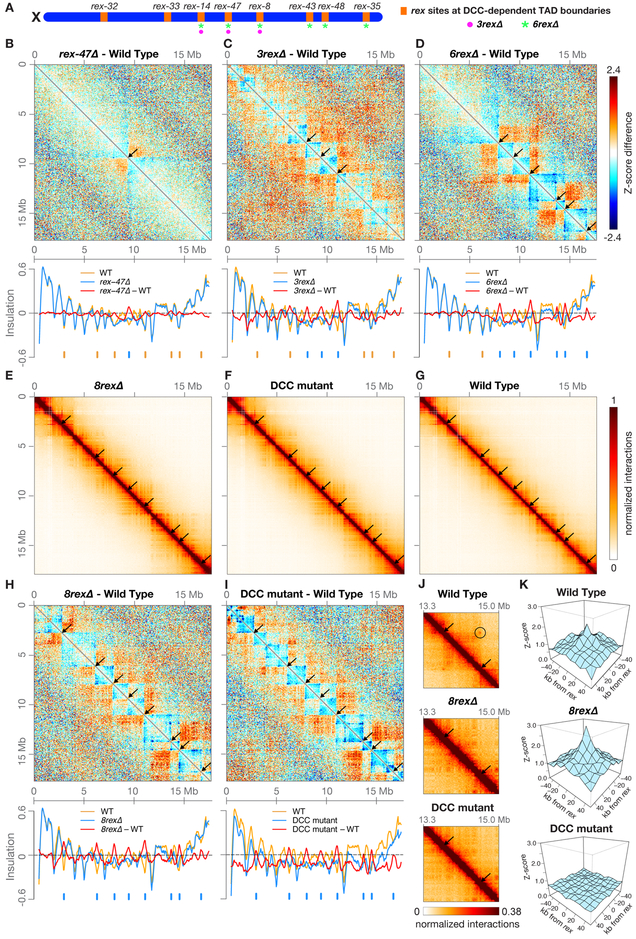
Figure 1. A Single rex Site Is Necessary for Formation of Each DCC-Dependent TAD Boundary.
(A) Locations on X of the eight rex sites (orange) at DCC-dependent TAD boundaries. All eight sites are deleted on 8rexΔ chromosomes, but only a subset are deleted on rex-47Δ, 3rexΔ (magenta circles), and 6rexΔ (green asterisks) chromosomes.
(B, C, and D) X chromosome heatmaps binned at 50 kb show wild-type Hi-C Z-scores subtracted from rex-47Δ, 3rexΔ, or 6rexΔ Hi-C Z-scores. Red, higher interactions in rex mutant. Blue, higher interactions in wild type. Arrows, positions of deleted rex sites where TAD boundaries are lost. Plots (below) show insulation scores across X in rex deletion mutants (blue) and wild-type embryos (orange) and the insulation score difference between genotypes (red). Blue ticks, positions of deleted rex sites. Orange ticks, DCC-dependent boundaries that persist in the mutant.
(E, F, and G) X heatmaps binned at 20 kb show Hi-C interactions in 8rexΔ, DCC mutant [sdc-2(y93, RNAi)], and wild-type embryos. Arrows mark positions of DCC-dependent boundaries found in wild-type embryos, which are lost in both mutants. Other DCC-independent boundaries remain.
(H and I) Heatmaps binned at 50 kb compare Hi-C Z-scores in 8rexΔ or DCC mutants to those in wild-type embryos. Plots (below) show X chromosome insulation profiles. Black arrows (top) and blue ticks (bottom) show locations of DCC-dependent boundaries.
(J) Heatmaps binned at 10 kb show enlargement of the X region surrounding rex-43 and rex-48 (arrows) in wild-type and mutant embryos and the interaction between the two rex sites in wild-type embryos (circle).
(K) 3D plots show that average Z-scores increase in 8rexΔ versus wild-type embryos for interactions among the 22 non-boundary rex sites with highest SDC-3 binding. Shown are interactions between sites within 4 Mb. DCC-mediated rex interactions occur regardless of the orientation of known X-enriched motifs (Jans et al., 2009) that are important in rex sites for DCC binding (Figure S3A). Insulation scores were calculated by summing interactions in a 500 kb sliding window.
See also Figures S1–S3, Tables S1 and S2.