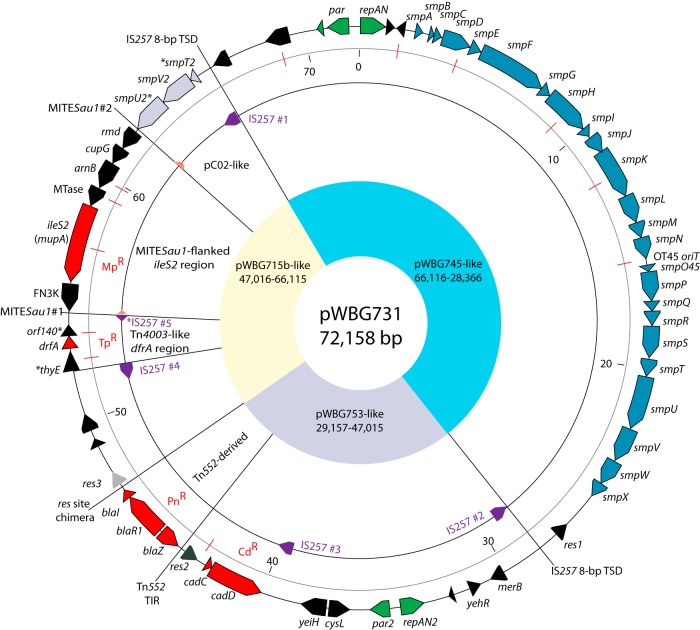
FIG 1.
The 72-kb conjugative multiresistance plasmid pWBG731. Map of the pWBG731 sequence highlighting sequence features. The outer ring shows positions of predicted genes and key cis-acting sequences likely involved in its evolutionary construction. Asterisks before or after a gene name indicate a 5′ or 3′ gene truncation, respectively. The gray ring shows the sequence ruler (in kilobase pairs), and positions of ClaI sites (corresponding to ClaI-digested DNA in Fig. 4B) are shown as red lines. The next inner ring highlights positions of IS257 and MITESau1 elements. All IS257 copies except IS257#5 carry an intact transposase gene. Yellow, blue, and gray sectors indicate regions nearly identical to those present on plasmids pWBG745, pWBG753, and pWBG715. TIR, terminal inverted repeat; MTase, predicted class I SAM-dependent methyltransferase domain-encoding gene.