Multidrug-resistant tuberculosis (TB) is an alarming threat, and targeted deep sequencing (DS) may be an effective method for rapid identification of drug-resistant profiles, including detection of heteroresistance. We evaluated the sensitivity and specificity of targeted DS versus phenotypic drug susceptibility testing (pDST) among patients starting first-line anti-TB therapy in Botswana. Overall, we found high concordance between DS and pDST.
KEYWORDS: HIV infections, next-generation sequencing, diagnostics, drug-resistant tuberculosis, heteroresistance, single-molecule overlapping reads
ABSTRACT
Multidrug-resistant tuberculosis (TB) is an alarming threat, and targeted deep sequencing (DS) may be an effective method for rapid identification of drug-resistant profiles, including detection of heteroresistance. We evaluated the sensitivity and specificity of targeted DS versus phenotypic drug susceptibility testing (pDST) among patients starting first-line anti-TB therapy in Botswana. Overall, we found high concordance between DS and pDST. Lower sensitivity of DS, which targets established high-confidence resistance variants, was observed for detecting isoniazid resistance among HIV-infected patients.
TEXT
The recent rollout of molecular-based diagnostic approaches represents an important step forward in reducing the time required to identify multidrug-resistant tuberculosis (MDR-TB; defined as resistance to both isoniazid [INH] and rifampin [RIF]) (1). However, widely used methods such as GeneXpert MTB/RIF and line-probe assays may not detect drug-resistant strains when they represent a small proportion of the population due to microevolution or concomitant infection with drug-susceptible strains (mixed infections) (2, 3). Targeted deep sequencing with the single-molecule-overlapping read (SMOR) assay is a promising molecular approach that allows rapid characterization of drug-resistant profiles by targeting resistance-conferring mutations at multiple bacterial loci with high levels of sequencing depth (e.g., >10,000× coverage) (4–6) and enables the identification and quantification of rare and ultrarare genetic variants down to a resolution as low as 0.1% in a heterogeneous sample (7).
We examined the clinical utility (sensitivity and specificity) of deep sequencing in detecting INH and RIF resistance at reported analytical sensitivities for detecting minority drug-resistant variants (resistance-associated variants [RAV]) for GeneXpert MTB/RIF (≥50%) (8), line-probe assays (≥5%) (2), and phenotypic drug susceptibility testing (pDST) with the mycobacteria growth indicator tube (MGIT; ≥1%) (2) among patients starting first-line anti-TB therapy in Botswana (1). From 2012 to 2016, the Kopanyo study enrolled all patients diagnosed with TB in the Gaborone and Ghanzi districts of Botswana (9). Mycobacterium tuberculosis DNA was extracted from positive sputum cultures of recruited patients and underwent 24-locus mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) genotyping.
Based on MIRU-VNTR, 99 patients with mixed or possibly mixed infections (defined as the presence of ≥2 repeats at one or more loci in the same sputum sample) and 200 patients randomly selected from 1,396 patients with single-strain infection were included in the present study. M. tuberculosis DNA from primary cultures used for MIRU-VNTR underwent targeted deep sequencing with the SMOR assay (5, 6). We targeted three critical gene regions known to confer resistance with a 25,000× depth of coverage, inhA promoter and the katG gene (associated with INH resistance) (10) and the RIF resistance-determining region (RRDR) of the rpoB gene (11). Fifteen samples with an average depth of <2,000 total reads were excluded to minimize misclassification of low-frequency variants due to sequencing errors (6). For the RIF specificity analysis, an additional 19 samples were excluded due to failed deep sequencing reads. pDST was performed for all cultured isolates with the MGIT 960 system and was used as the reference standard for sensitivity and specificity calculations. All discordant samples between deep sequencing and pDST were retested with MGIT.
Overall, 241 patients with known HIV status and pDST results were included in the final analysis (Table 1). Of these, 222 (92.1%) patients had M. tuberculosis strains that were susceptible to both drugs, 8 (3.3%) were monoresistant to INH, 4 (1.7%) were monoresistant to RIF, and 7 (2.9%) were resistant to both drugs (MDR-TB).
TABLE 1.
Characteristics of participantsa
| Characteristic | No. of HIV-negative patients (%) or median (IQR) | No. of HIV-positive patients (%) or median (IQR) | Allb patients (%) or median (IQR) |
|---|---|---|---|
| District | |||
| Gaborone | 79 (73.8) | 122 (91.0) | 207 (83.5) |
| Ghanzi | 28 (26.2) | 12 (9.0) | 41 (16.5) |
| Gender | |||
| Male | 63 (58.9) | 78 (58.2) | 145 (58.5) |
| Female | 44 (41.1) | 56 (41.8) | 103 (41.5) |
| Age in years | 28.4 (22.0–38.0) | 36.7 (32.6–44.0) | 34.2 (26.4–42.0) |
| Prior hospitalization | |||
| No | 64 (59.8) | 70 (52.2) | 139 (56.0) |
| Yes | 24 (22.4) | 40 (29.9) | 66 (26.6) |
| Unknown | 19 (17.8) | 24 (17.9) | 43 (17.3) |
| Prior TB | |||
| No | 95 (88.8) | 110 (82.1) | 211 (85.1) |
| Yes | 12 (11.2) | 24 (17.9) | 37 (14.9) |
| Smear microscopy | |||
| Negative | 12 (11.2) | 31 (23.1) | 46 (18.5) |
| Positive | 95 (88.8) | 103 (76.9) | 202 (81.5) |
| Phenotypic DST | |||
| Susceptible | 102 (95.3) | 120 (89.6) | 229 (92.3) |
| INH-monoresistant | 2 (1.9) | 6 (4.5) | 8 (3.2) |
| RIF-monoresistant | 0 | 4 (3.0) | 4 (1.6) |
| MDR-TB | 3 (2.8) | 4 (3.0) | 7 (2.8) |
| CD4+ T cell count | NA | 196 (104–394) | NA |
IQR, interquartile range; DST, drug susceptibility testing; INH, isoniazid; RIF, rifampin; MDR-TB, multidrug-resistant TB; NA, not available.
Includes 7 patients with unknown HIV status.
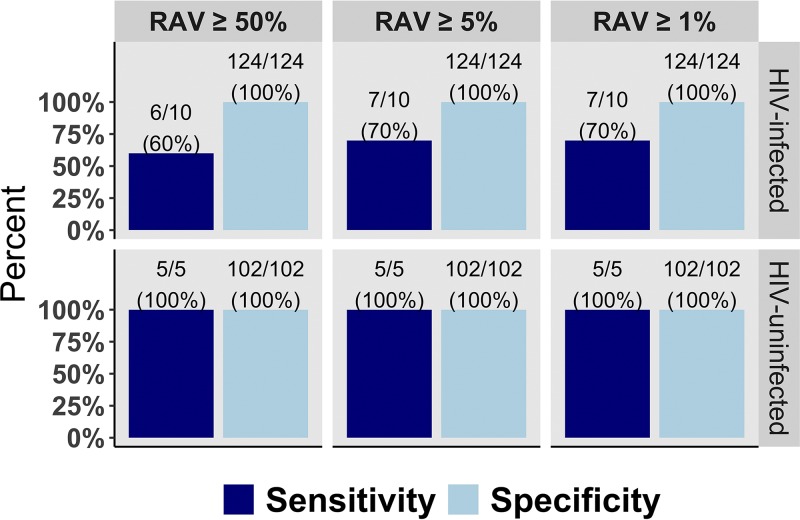
Deep sequencing of the targeted loci correctly identified those who were phenotypically susceptible to RIF (211/211; 100% specificity) and to INH (226/226; 100% specificity), regardless of HIV infection status (Fig. 1). Among 11 specimens that showed phenotypic resistance to RIF, deep sequencing identified all 11 as genotypically resistant (11/11; 100%), regardless of HIV infection status. Among HIV-infected patients, targeted deep sequencing detected 6 out of 10 patients with phenotypic INH resistance at RAV frequency of ≥50% and 7 out of 10 patients at RAV frequency of ≥5% and ≥1% (versus 100% among HIV-uninfected patients; Fig. 1). The overall sensitivity of deep sequencing for INH resistance detection (regardless of HIV infection status) was 80% (12/15) at RAV frequency of ≥1%. All INH-resistant isolates had a MIC of 0.2 μg/ml. No differences were found between isolates with different RAV frequencies. We verified all discordant samples at RAV frequency of ≥5% via SMOR and confirmed that there were no minor variants or any other single-nucleotide polymorphisms (SNPs) occurring within the inhA promoter and katG high-confidence resistance regions.
FIG 1.
Sensitivity and specificity of targeted deep sequencing in detecting INH resistance at different RAV frequencies, stratified by HIV infection status. INH, isoniazid; RAV, resistance-associated variants.
Table 2 shows the characteristics of patients with discordance between deep sequencing and pDST at RAV frequency of ≥50%. All four discordant cases were HIV positive; one had single-strain infection, two had possibly mixed infections, and one had mixed infection with 17% RAV detected in the katG gene.
TABLE 2.
Characteristics of patients with discordance between deep sequencing and pDST at RAV frequency of ≥50%a
| Patient | Gender | Age (yr) | HIV | CD4+ T cell count | Smear microscopy | pDST INH | pDST RIF | MIRU-VNTR | TB treatment outcome | RAV frequency (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 47 | Positive | 142 | Positive | Resistant | Susceptible | Single strain | Defaulted | <0.1 all loci |
| 2 | Male | 28 | Positive | Unknown | Positive | Resistant | Susceptible | Possibly mixed infectionb | Completed | <0.1 all loci |
| 3 | Female | 34 | Positive | 773 | Positive | Resistant | Susceptible | Possibly mixed infectionb | Completed | <0.1 all loci |
| 4 | Male | 39 | Positive | 17 | Negative | Resistant | Susceptible | Mixed infectionc | Completed | 17.01 katG 315ACC |
Abbreviations: pDST, phenotypic drug susceptibility testing; INH, isoniazid; RIF, rifampin; MIRU-VNTR, 24-locus mycobacterial interspersed repetitive units-variable number of tandem repeats; RAV, resistance-associated variants.
Defined as the presence of ≥2 repeats at a single locus in the same sputum sample.
Defined as the presence of ≥2 repeats at more than one locus in the same sputum sample.
Overall, we found a very high concordance between deep sequencing and pDST among patients starting first-line TB therapy in Botswana. Sensitivity of deep sequencing of the targeted loci may be lower for detecting INH resistance among HIV-infected individuals. Notably, we were able to detect an INH-resistant isolate with 17% RAV in the katG gene, which could have been missed by less sensitive tests. This indicates that deep sequencing could, in principle, improve early detection of INH resistance compared to conventional molecular tests in clinical settings where there is a high prevalence of heteroresistant infections.
Our findings are consistent with studies by other groups that observed reduced sensitivity for molecular detection of INH resistance in high-HIV settings. Dorman et al. reported 62% sensitivity for MTBDRplus v1 in detecting INH resistance among TB patients in South Africa (12), while Luetkemeyer et al. reported 70.6% sensitivity among HIV-infected patients in South Africa, Botswana, and South America (13). It is possible that the mechanisms of acquiring resistance are different in some HIV-positive patients who were previously treated with INH preventive therapy (14).
Our findings also highlight the possibility that noncanonical mutations not covered by our SMOR assay may be responsible for clinical INH resistance, particularly among HIV-infected TB patients. While katG and inhA promoter mutations explain the majority of resistance to INH, a subset of INH-resistant clinical isolates do not carry these common mutations (15). Mutations in katG and inhA promoter genes have been shown to confer low fitness costs, allowing the mutant microbe to survive and propagate without negative selection pressure (16). It is possible that mutant M. tuberculosis strains with reduced fitness are more likely to survive and replicate in immunocompromised HIV-infected hosts, and these mutations may be missed by targeting only high-confidence, well-published RAVs. However, our results are limited by the small sample size of patients with drug-resistant TB. Additional research on a larger sample size of drug-resistant TB patients is needed to confirm our findings.
ACKNOWLEDGMENTS
We are thankful to the study participants and the Kopanyo study team, who made this work possible.
This study was funded by the National Institutes of Health (grants R01AI097045, P30AI45008, K01AI118559, and R21AI120838). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.World Health Organization. 2018. Global tuberculosis report 2018. https://www.who.int/tb/publications/global_report/en/.
- 2.Folkvardsen DB, Svensson E, Thomsen VO, Rasmussen EM, Bang D, Werngren J, Hoffner S, Hillemann D, Rigouts L. 2013. Can molecular methods detect 1% isoniazid resistance in Mycobacterium tuberculosis? J Clin Microbiol 51:1596–1599. doi: 10.1128/JCM.00472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinder H, Mieskes KT, Loscher T. 2001. Heteroresistance in Mycobacterium tuberculosis. Int J Tuber Lung Dis 5:339–345. [PubMed] [Google Scholar]
- 4.Metcalfe JZ, Streicher E, Theron G, Colman RE, Allender C, Lemmer D, Warren R, Engelthaler DM. 2017. Cryptic microheteroresistance explains Mycobacterium tuberculosis phenotypic resistance. Am J Respir Crit Care Med 196:1191–1201. doi: 10.1164/rccm.201703-0556OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman RE, Anderson J, Lemmer D, Lehmkuhl E, Georghiou SB, Heaton H, Wiggins K, Gillece JD, Schupp JM, Catanzaro DG, Crudu V, Cohen T, Rodwell TC, Engelthaler DM. 2016. Rapid drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates directly from clinical samples by use of amplicon sequencing: a proof-of-concept study. J Clin Microbiol 54:2058–2067. doi: 10.1128/JCM.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, Valafar F, Crudu V, Romancenco E, Noroc E, Jackson L, Catanzaro DG, Rodwell TC, Catanzaro A, Keim P, Engelthaler DM. 2015. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLoS One 10:e0126626. doi: 10.1371/journal.pone.0126626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. 2012. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol 48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zetola NM, Modongo C, Moonan PK, Click E, Oeltmann JE, Shepherd J, Finlay A. 2016. Protocol for a population-based molecular epidemiology study of tuberculosis transmission in a high HIV-burden setting: the Botswana Kopanyo study. BMJ Open 6:e010046. doi: 10.1136/bmjopen-2015-010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. 2015. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One 10:e0119628. doi: 10.1371/journal.pone.0119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, Matter L, Schopfer K, Bodmer T. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 12.Dorman SE, Chihota VN, Lewis JJ, van der Meulen M, Mathema B, Beylis N, Fielding KL, Grant AD, Churchyard GJ. 2012. Genotype MTBDRplus for direct detection of Mycobacterium tuberculosis and drug resistance in strains from gold miners in South Africa. J Clin Microbiol 50:1189–1194. doi: 10.1128/JCM.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luetkemeyer AF, Kendall MA, Wu X, Lourenco MC, Jentsch U, Swindells S, Qasba SS, Sanchez J, Havlir DV, Grinsztejn B, Sanne IM, Firnhaber C, Adult ACTGAST. 2014. Evaluation of two line probe assays for rapid detection of Mycobacterium tuberculosis, tuberculosis (TB) drug resistance, and non-TB Mycobacteria in HIV-infected individuals with suspected TB. J Clin Microbiol 52:1052–1059. doi: 10.1128/JCM.02639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2011. Intensified tuberculosis case-finding and isoniazid preventive therapy for people living with HIV in resource-constrained settings: WHO guidelines 2011. https://www.who.int/hiv/pub/tb/9789241500708/en/.
- 15.Kandler JL, Mercante AD, Dalton TL, Ezewudo MN, Cowan LS, Burns SP, Metchock B, Global PETTS Investigators, Cegielski P, Posey JE. 2018. Validation of novel Mycobacterium tuberculosis isoniazid resistance mutations not detectable by common molecular tests. Antimicrob Agents Chemother 62:e00974-18. doi: 10.1128/AAC.00974-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borrell S, Gagneux S. 2009. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuber Lung Dis 13:1456–1466. [PubMed] [Google Scholar]