Figure 3.
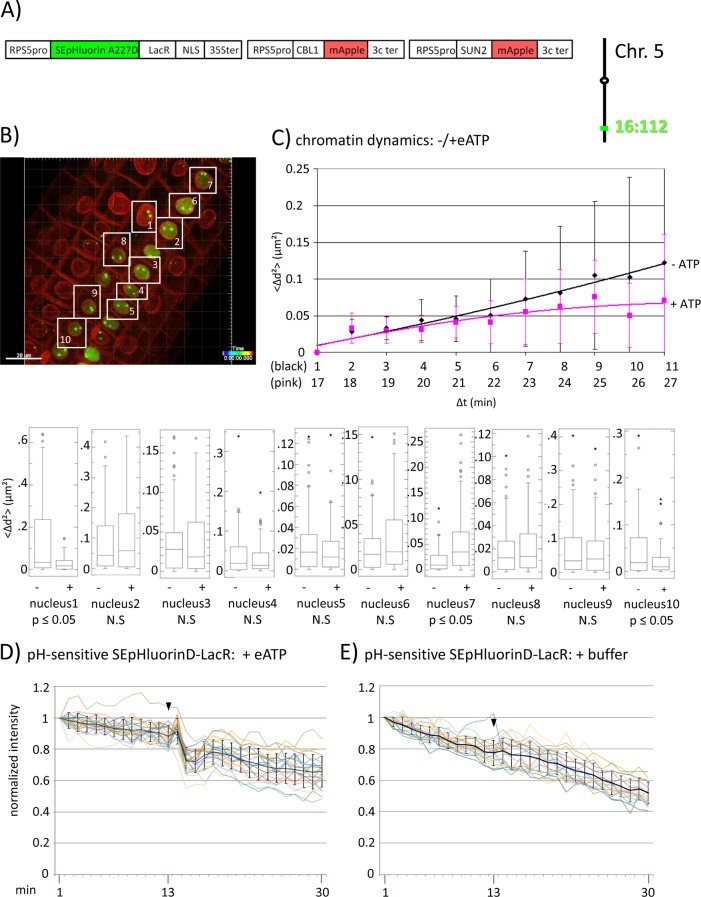
Fluorescence intensity changes of a pH-sensitive fluorescent DNA-binding protein following addition of eATP. (A) The construct encoded nuclear-localized SEpHluorinD-LacR fusion protein as a pH-sensitive fluorescent DNA-binding protein, and SUN2-mApple and CBL1-mApple as INM and PM visual markers. All three genes are under the transcriptional control of the RPS5 promoter; the first contains the 35ter and the last two contain the 3C terminator (3Cter) for readability, the module encoding RPS5pro-LacR is in the orientation shown but it is actually in the opposite orientation (Data Sheet 1, combination Figure 3, in supplement). Right: The SEpHluorinD-LacR fusion protein binds to lacO repeats integrated at locus 16:112 on the bottom arm of chromosome 5. (B) Confocal image (maximum projection; enlargement in Data Sheet 9 in supplement) at time point t1 of homozygous fluorescent-tagged locus 16:112 in nuclei of cells in the root transition zone. Two green dots are visible in most nuclei. Nuclei used for chromatin dynamics analyses (part C) and fluorescence intensity analysis (parts D and E) are boxed in white. (C) Chromatin dynamics: Mounting, confocal microscopy, eATP treatment, data acquisition and data analysis of Arabidopsis seedlings harboring the above construct were carried out as described in the legend to Figure 2. Chromatin dynamics was determined as described in the legend to Figure 1. The graph shows a scatterplot of the average Δd2 (<Δd2>) values [cumulative (squared) distance travel since t0 and after eATP addition since t17], and standard deviation bars (Table 2, sheet 6 in supplement) for 10 nuclei over a period of 30 min (Δt = elapsed time since t0) and is overlaid with order 2 polynomial trendlines, which indicate “jiggling” before (black) and after (pink) eATP addition. Time points 12-16 were excluded from the analysis owing to unreliable data acquired during dislocation turbulence caused by addition of eATP. To detect differences in the mobility of locus 16:112 before and after eATP treatment, the analysis was restarted following the addition of eATP, hence producing two lines. Bottom: Single nuclei box plot analyses and calculation of p values using the Δd2 values revealed three nuclei with significantly different changes (p ≤ 0.05) in the mobility of locus 16:112 following eATP treatment. N.S., not significant. (D) pH-sensitive SEpHluorin-LacR chromatin tag: normalized fluorescence intensity profiles of the two 16:112 alleles in the 10 white-boxed nuclei (numbered in white in part B) are overlaid in one graph together with the calculated average values, to which standard deviation bars were added. eATP addition is indicated with a black arrowhead at frames 13-14. Fluorescence intensities of individual nuclei are shown in supplementary Data Sheet 8, part 3D, in which the boxed areas in the fluorescent intensity graphs highlight the region of interest. ΔpH values are shown under each nucleus1-10 for both alleles [maximum ΔpH 0.4 (nucleus 9); minimum ΔpH 0.1 (nuclei 4 and 5); average (n = 20) 0.2]. Normalized and non-normalized data are shown, respectively, in Data Sheets 3 of Tables 10 and 1. Using the normalized data, the calculated difference in the magnitude of the drop in fluorescence intensity in SEpHluorinD-LacR plus eATP versus bleaching in the buffer control between the time points 14-16 points is statistically significant (p ≤ 0.05). (E) pH-sensitive SEpHluorinD-LacR chromatin tag: Buffer control without eATP (original data can be viewed in supplementary Table 1, sheet 4). The spikes in fluorescence reflect dislocation turbulence, which occurs upon addition of eATP or buffer. The fluorescence intensity at the genomic location is read in spots objects (1-2.8 μm) capturing punctual fluorescence of the tagged regions.