Abstract
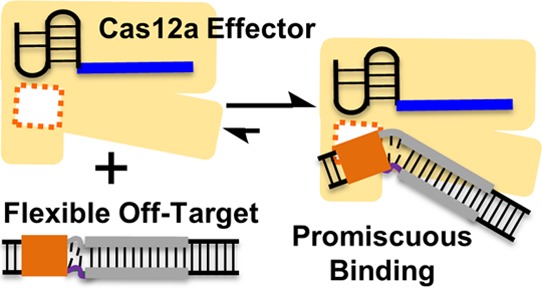
Cas12a (also known as “Cpf1”) is a class 2 type V-A CRISPR-associated nuclease that can cleave double-stranded DNA at specific sites. The Cas12a effector enzyme comprises a single protein and a CRISPR-encoded small RNA (crRNA) and has been used for genome editing and manipulation. Work reported here examined in vitro interactions between the Cas12a effector enzyme and DNA duplexes with varying states of base-pairing between the two strands. The data revealed that in the absence of complementarity between the crRNA guide and the DNA target-strand, Cas12a binds duplexes with unpaired segments. These off-target duplexes were bound at the Cas12a site responsible for RNA-guided double-stranded DNA binding but were not cleaved due to the lack of RNA/DNA hybrid formation. Such promiscuous binding was attributed to increased DNA flexibility induced by the unpaired segment present next to the protospacer-adjacent-motif. The results suggest that target discrimination of Cas12a can be influenced by flexibility of the DNA. As such, in addition to the linear sequence, flexibility and other physical properties of the DNA should be considered in Cas12a-based genome engineering applications.
Introduction
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) proteins constitute a class of adaptive immune system used by bacteria and archaea.1 A number of CRISPR-Cas systems have been adapted for genome engineering,2 serving as programmable nucleases cleaving desired genomic sequences3−6 or as specific DNA binders enabling transcriptional regulation,7,8 base editing,9−11 or imaging.12 Among them, Cas12a (also known as “Cpf1”) is a class 2 type V CRISPR that has been used for gene editing,6,13−15 transcriptional regulation,16−19 and base-editing.20 These applications rely on a binary effector complex formed by a single Cas12a protein and CRISPR-encoded small RNA (crRNA). A Cas12a effector cleaves double-stranded DNAs that meet two criteria: (a) base complementarity between the crRNA guide region and a segment of the DNA duplex designated as the “protospacer” and (b) a short DNA motif flanking the protospacer, designated as the protospacer-adjacent-motif (PAM).6 Since its discovery in 2015, rapid progress is being made in delineating Cas12a mechanisms. High-resolution structures have been reported for Cas12a complexes.21−29 Mechanistic understanding is emerging on crRNA activation,21,25 PAM recognition,22,24,27 duplex unwinding to form an R-loop,22,23,25,26,28−32 and R-loop dependent DNA cleavage.26,28,30−32 The studies have revealed that similar to the better-studied Cas9, Cas12a uses nucleic-acid dependent conformational changes as checkpoints to achieve cleavage of correct DNA targets, although details of the Cas12a checkpoints have distinct features.21,22,25,26,28,30−32 Furthermore, in addition to crRNA-guide dependent cleavage of double-stranded DNAs (i.e., the cis-activity),6,29 Cas12a can carry out other enzymatic functions. For example, in the presence of certain divalent metal ions, the apo-Cas12a can cleave DNA duplex in a crRNA-independent manner.33 Studies have also uncovered a “trans-activity” of Cas12a, referring to nonspecific cleavage of single-stranded DNAs by an “activated” ternary Cas12a-crRNA-DNA complex.28,29,34,35 However, much remains to be learned about mechanisms of Cas12a.
Studies on mechanisms of CRISPR-Cas, including Cas12a, is essential not only for developing applications targeting various forms of DNA3−6,34 or RNA,36−38 but also for understanding and minimizing interactions with undesired targets. In a majority of Cas9- and Cas12a-based genome engineering applications, the correct DNA target (“on-target”) presents full complementarity between the RNA-guide and the target-strand of the DNA protospacer. A major class of the undesired targets are off-target DNAs that have mismatch(es) between the RNA-guide and the DNA target-strand.39−41 Off-target effects have been recognized as a critical issue in genome engineering: off-target cleavage results in gene disruption at undesired locations,39−41 whereas off-target binding interferes with CRISPR-based imaging,42,43 transcriptional regulation, and base-editing.44,45 Mechanistic understanding46 has been playing a prominent role in combating off-target effects and has led to successful developments of high fidelity Cas947−49 and Cas12a50 variants.
Current studies on target discrimination by Cas9 and Cas12a have primarily focused on the protein-RNA complex,47−52 with much less attention being paid on how the physical property of the DNA may impact CRISPR target selection.53 However, a large body of work on DNA-protein and DNA-small-molecule interaction has firmly established the importance of DNA physical properties in determining specificity.54 For example, in DNA recognition by transcription factors, specificity is derived from two modes that contribute nearly equally:54 (a) base-readout, in which the protein recognizes DNA base functional groups; and (b) shape-readout, in which the protein recognizes collective physical properties of the duplex (e.g., flexibility, minor groove width, and local electrostatics). Indeed, there is evidence that physical properties of the DNA are utilized in CRISPR target recognition, for example, PAM recognition has been found to involve not only base-readout55 but also shape-readout.22
Structures of ternary Cas946 and Cas12a22,28 complexes show that the bound DNA is severely kinked at the junction between the PAM and the protospacer. In addition, for Cas9 it has been shown that DNA distortion occurs prior to duplex unwinding and R-loop formation.56 As such, DNA flexibility, the major factor dictating how easy a DNA can be distorted, may impact Cas9 and Cas12a target recognition. Other than its sequence, flexibility of a given DNA can alter significantly due to a variety of factors. For example, during transcription or replication, the double-stranded DNAs contain unpaired segments (i.e., “bubbles”) with significantly higher flexibility. Indeed, Rueda and co-workers recently reported that DNA bubbles increase Cas9 off-target binding and cleavage.53 However, detailed studies have not been reported on how DNA flexibility may impact Cas12a off-target recognition, although a number of recent publications have used duplexes that contain bubbles to study mechanisms of Cas12a.28,29,32
In this report, we examined in vitro Cas12a interactions with DNAs whose protospacer contained “bubble(s)”, that is, stretch(es) of unpaired nucleotides between the target-strand (i.e., “t-strand”) and nontarget strand (“nt-strand”). The data revealed that in the absence of complementarity between the DNA protospacer t-strand and the crRNA guide, PAM-adjacent bubbles allowed duplex binding but not cleavage. Such binding occurred at the same site used by Cas12a for RNA-guided binding of on-target double-stranded DNAs, and was attributed to increased DNA flexibility induced by the bubble. The results suggest that DNA binding of Cas12a can be influenced by flexibility of the DNA duplex.
Results
Reconstructing Cas12a Systems for in Vitro Characterizations
DNA recognition was studied using recombinant Cas12a proteins from Lachnospiraceae bacterium (LbCas12a), Acidaminococcus sp. BV3L6 (AsCas12a), and Francisella novicida (FnCas12a).6,33 The corresponding crRNAs were named according to its respective ortholog-specific repeat sequence and the guide sequence, for example, LbRNA-a represented a crRNA with the repeat sequence of Lachnospiraceae bacterium and the RNA-a guide sequence [Figure 1A, Supporting Information Section S1]. The DNA substrate duplexes contained a 5′-TTTC-3′ PAM flanking a 20-nucleotide protospacer (Figure 1A; Supporting Information Section S2). The reconstituted binary Cas12a/crRNA enzymes, as expected, cleaved the fully-paired duplex i that maintained perfect complementarity between the RNA-a guide and the protospacer t-strand (Figure 1, Supporting Information Section S3). Control experiments showed that the cognate substrate was cleaved at the proper sites (Supporting Information Section S3), and the activities required crRNA (Supporting Information Section S4) but were independent of the presence of the maltose-binding-protein tag (Supporting Information Section S5).
Figure 1.

Reconstituting the Cas12a enzyme. (A) Schematic of a Cas12a/RNA/DNA construct, with the RNA-a guide sequence shown in blue and the red wedges marking the DNA cleavage sites (see Supporting Information Sections S1–S3 for more details). (B) Schematics of DNA duplex substrates (see also Table S3), with “R/D” indicating complementarity between the RNA guide and the t-strand of the DNA protospacer, and “D/D” indicating pairing between DNA t- and nt-strand. (C,D) Representative gels of cleavage of duplexes i–iv (10 nM each) by effector complex (100 nM) of LbCas12a/LbRNA-a or AsCas12a/AsRNA-a, respectively. Reactions were monitored using 5′-32P labeled t-strand, with precursors marked by “■” and t-strand products marked by “▶”.
Experiments were then carried out for duplexes with or without bubbles in the protospacer (Figure 1, Supporting Information Table S3). Independent of the presence of bubbles, cleavages were observed for “on-target” duplexes that maintained t-strand complementarity to the RNA-a guide (Figure 1, duplexes i & ii; Supporting Information Section S6) but were absent for “off-targets” that lacked RNA-a guide/t-strand complementarity (Figure 1, duplexes iii & iv; Supporting Information Section S6). Similar results were observed with a different RNA guide sequence (Supporting Information Section S7). Note that cleavage of the bubble-containing duplexes ii gave cryptic t-strand products (Figure 1C,D). This is similar to results reported by Swarts and Jinek29 and indicates that the bubble at the protospacer impacts how the Cas12a nuclease engages the DNA. In addition, a lack of cleavage for bubble-containing duplex iii (Figure 1, Supporting Information Section S6) is consistent with a recent report showing that only 2 or 4 RNA/DNA mismatches at the PAM-adjacent positions prevent cleavage of bubble duplexes.32
A bubble in the protospacer may be considered as “single-stranded” substrates. However, with the Cas12a enzyme containing the RNA-a guide, the “on-target” duplex ii gave specific t- and nt-strand products, whereas the off-target duplex iii had no cleavage (Figure 1C,D, Supporting Information Section S6). This indicates that under the single-turnover condition used in this work, the results reported the double-stranded DNA cleavage activity of the binary Cas12a/crRNA effector (i.e., the cis-activity), and did not reflect the nonspecific trans-activity on single-stranded DNAs. This is indeed confirmed in control experiments (Supporting Information Section S8).
Protospacer Flexibility Promotes Promiscuous Off-Target Binding to Cas12a
A competition assay57 was adapted to examine DNA duplex binding by Cas12a. In these experiments (Figure 2A), cleavage of a 32P-labeled duplex (i.e., “probe”) by an effector was measured under single-turnover conditions in the presence of an unlabeled DNA competitor. Reactions were carried out with [probe] ≪ [enzyme] ≪ [competitor], and the probe and competitor were presented simultaneously to the enzyme (see Materials and Methods). If a competitor binds to the same enzyme site as that of the probe, probe binding is inhibited, thus reducing or preventing probe cleavage. Because the observed single-turnover cleavage of the probe reported only on the cis-activity of the Cas12a/crRNA effector (Supporting Information Section S8), the competition experiment assesses DNA binding at the site responsible for RNA-guide dependent binding and cleavage of double-stranded DNAs.
Figure 2.

Assessing DNA binding to Cas12a. (A) Schematic of the competition assay. (B) Schematics of the competitor duplexes (see also Table S3). (C,D) Representative gels of competitions with LbCas12a/LbRNA-a and AsCas12a/AsRNA-a, respectively. Data shown were obtained with 1 nM probe i* (32P-labeled t-strand), 10 nM enzyme, and 1 μM competitors.
Competitions were first carried out with 32P-labeled duplex i* as the “probe” and unlabeled duplexes i to iv as competitors (Figure 2B). The enzyme is the LbCas12a/LbRNA-a complex, which contained the RNA-a guide complementing the t-strand of the protospacer of i*. Figure 2C shows a representative set of data obtained with 10 nM enzyme and 1 μM competitors. Duplexes i and ii, which maintain protospacer t-strand complementarity to the LbRNA-a guide (Figure 2B), inhibited cleavage of i* (Figure 2C, Supporting Information Section S9a, Table S4). This indicates that as expected both duplexes bind to the LbCas12a active site and is consistent with results from the cleavage assay (Figure 1). Interestingly, duplexes iii and iv gave clearly different results even though both largely lack RNA-guide/t-strand complementarity and were not cleaved in the cleavage assay (Figure 1C). Duplex iv showed little observable inhibition (Figure 2C, Table S4), indicating that it did not bind to Cas12a. On the other hand, duplex iii significantly reduced probe i* cleavage, indicating sufficient binding to the Cas12a to exert inhibition (Figure 2C, Table S4). Inhibition experiments with AsCas12a and FnCas12a yielded an identical pattern (Figure 2D, Supporting Information Section 9b) and similar results were obtained at a higher concentration (100 nM) of enzyme effector complexes (Supporting Information, Section S9c). Overall the competition data revealed that for Cas12a effectors containing the RNA-a guide, the bubble-containing duplex iii binds significantly better than the fully paired duplex iv. This is further confirmed by direct measurements of DNA binding (Supporting Information Section S10).
Neither duplex iii nor iv allowed hybrid formation between the RNA-a guide and the protospacer t-strand. The difference in their inhibition of (and binding to) RNA-a containing Cas12a effectors (Figure 2C,D, Table S4) must arise from intrinsic physical properties of the DNA itself—the protospacer maintains perfect t-/nt-strand pairing in duplex iv but is largely unpaired in duplex iii (Figure 2B). An unpaired segment (i.e., bubble) within a DNA duplex is significantly more flexible than a fully paired segment.58 Indeed, using a biophysical technique called site-directed spin labeling,59 duplex iii was found to be significantly more flexible than that of a fully-paired duplex (Supporting Information Section S11). In addition, control experiments showed that the proper PAM in duplex iii contributed to Cas12a binding (Supporting Information Section S9d). Taken together, the data support a model that upon PAM-binding, the more-flexible duplex iii can distort to remain bound to Cas12a, thus blocking on-target duplex (i.e., i*) binding and preventing cleavage (see Discussion). On the other hand, even with a proper PAM, the rigid fully paired duplex iv cannot distort sufficiently to stay bound to Cas12a (see Discussion), therefore exhibiting little inhibition. The effect of the protospacer flexibility would be independent of the sequence of the RNA guide, as indeed demonstrated by control experiments (Supporting Information, Section S9e).
PAM-Adjacent Flexibility Enables Promiscuous Binding
To further examine the relationship between protospacer flexibility and off-target Cas12a binding, duplex iii variants were constructed. No measurable difference was observed when the PAM-distal nonconsecutive complementary nucleotides in duplex iii were removed (Supporting Information Section S9f), indicating that further enlarging the protospacer bubble has little impact on Cas12a binding. Studies were then carried out with off-target duplexes containing PAM-adjacent protospacer bubbles ranging from 1 to 4 base-pairs (bp) (duplexes v to viii, Figure 3A, Table S3). Competition showed that duplex viii effectively inhibited i* cleavage by a Cas12a complex containing a RNA-a guide (Figure 3, Table S5). The degree of inhibition observed for viii, which has a 4 bp PAM-adjacent bubble, was similar to that obtained with duplex iii, which has the bubble spanning the entire protospacer (Table S5). This indicates that a PAM-adjacent bubble of 4 bp was sufficient to elicit promiscuous binding to Cas12a in the absence of RNA/DNA complementarity. As the size of the bubble decreased from 4 to 0 bp, the amount of probe cleavage increased monotonically (Figure 3, Table S5). This indicates a reduction of inhibition, which was attributed to a weaker binding of the competitor to the Cas12a complex. As the PAM-adjacent bubble was reduced in size, the DNA became less flexible and more difficult to distort, and this correlated with a decrease in affinity to Cas12a (i.e., less effective inhibition). This supports the notion that flexibility at the PAM-adjacent protospacer segment allows Cas12a binding of off-target duplexes.
Figure 3.
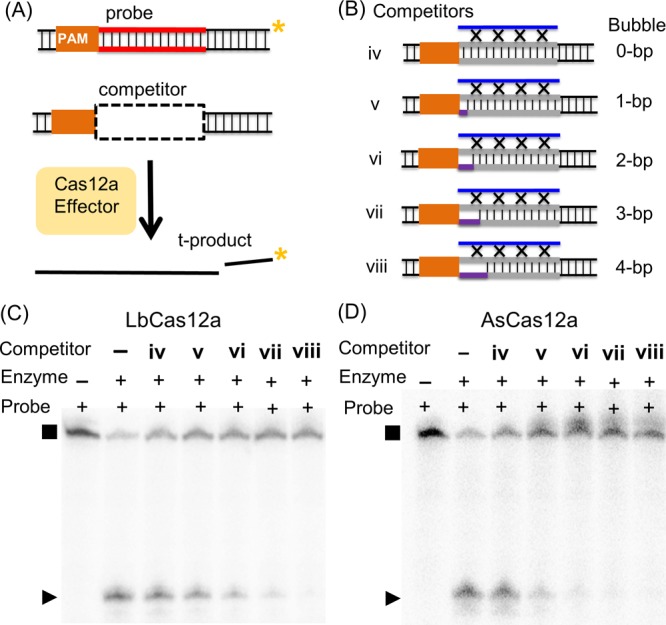
Assessing binding to Cas12a by DNA with PAM-adjacent bubbles. Shown in (A) is a schematic of the competition assay. Shown in (B) are schematics of the competitor duplexes (see also Table S3). Shown in (C,D) are representative gels of competitions with LbCas12a/LbRNA-a and AsCas12a/AsRNA-a, respectively. Data shown were obtained with 1 nM probe i* (32P-labeled t-strand), 10 nM enzyme, and 1 μM competitors.
Also note that as the bubble size increased, AsCas12a gave a larger degree of reduction of i* product (Figure 3C,D, Table S5) compared to LbCas12a. This may suggest differences between Cas12a orthologs in binding flexible duplexes. This will be an interesting subject for future investigations.
Discussion
Data presented in this work support the conclusion that flexibility at the PAM-adjacent protospacer segment enables DNA binding by Cas12a in the absence of DNA/RNA complementarity. The results are consistent with the current Cas12a mechanistic model and expand the understanding on how the physical property of the DNA may influence Cas12a target discrimination.
Current literature22,25,26,30 support a mechanistic model that Cas12a selects its duplexed DNA substrate via (i) PAM binding; (ii) PAM-adjacent protospacer distortion to initiate DNA unwinding; (iii) propagation of DNA unwinding with concomitant RNA/DNA hybrid base-pairing; and (iv) strand cleavage (Figure 4A). With PAM-containing off-target DNA duplexes that lack complementarity between the RNA-guide and DNA protospacer (e.g., RNA-a guide with duplexes iii to viii), the difference in Cas12a binding must lie at step (ii), that is, PAM-adjacent protospacer distortion. For these off-target duplexes, PAM-adjacent local melting also would expose the first few t-strand nucleotides, but the lack of RNA-guide/t-strand complementarity does not support RNA-DNA hybridization. The DNA pairing state then significantly influences the outcome as demonstrated by data presented here. With a fully-paired protospacer (i.e., duplex iv) (Figure 4B), the initially unwound nucleotides likely snap back to regain t-/nt-strand pairings. The paired duplex is rigid, and the junction between the PAM and the protospacer segment disfavors the kinked conformation as shown in the ternary Cas12a-RNA-DNA structures.22,23,25,26 This causes DNA dissociation as the protospacer may collide with the Cas12a effector. As such, a fully paired duplex predominantly favors the unbound state (Figure 4B), as observed for duplex iv (Figure 2, Supporting Information Section 10). On the other hand, for DNAs with a PAM-adjacent bubble (Figure 4C), flexibility at the PAM-protospacer junction increases. When sufficient flexibility exists (e.g. the 4 bp bubble duplex viii, Figure 3), the duplex can distort to assume conformations more similar to the kinked configurations observed in the ternary complexes22,23,25,26 (Figure 4C). The protospacer likely bends away to mitigate steric collision between the DNA and the effector complex, allowing a significant degree of binding even though DNA/RNA hybrid formation is not supported (Figure 4C).
Figure 4.
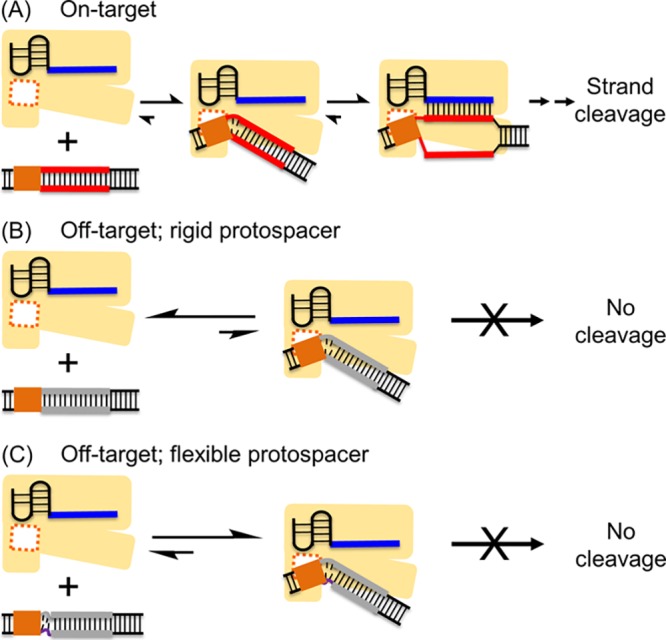
Models of DNA duplexes interacting with the binary Cas12a/crRNA effector enzyme. (A) On-target duplex. PAM binding is followed by local protospacer melting. Hybridization between the RNA guide and protospacer t-strand supports DNA unwinding and formation of the R-loop. (B) Off-target with a fully paired protospacer. Following PAM-binding, local protospacer melting can occur, but without the complementarity between the RNA guide and the protospacer t-strand, the DNA prefers to maintain the t- and nt-strand pairing, and cannot sustain the kinked configuration required to remain associated with the effector complex. As such, a paired (rigid) off-target duplex remains predominantly in the unbound state (left). (C) Off-target with PAM-adjacent protospacer bubble. When sufficient flexibility is provided by the bubble, the duplex can adapt a kinked configuration to remain bound to Cas12a (right). However, the RNA/DNA hybrid is absent; therefore, no cleavage occurs.
While the flexible off-target duplexes (e.g., iii and viii) bind to Cas12a, no cleavage was observed (Figure 1). Recent studies have revealed that RNA/DNA hybrid formation triggers the opening of the catalytic cleft,28 subsequently activating the cis- and then the trans-cleavage activities.28,29,34,35 In our present work, the off-target DNA duplexes, either with or without a bubble, do not support hybridization between the DNA and RNA guide (Table S3), thus accounting for their lack of cleavages either in the cis- or trans-fashion (Figure 1 and Supporting Information Section S8). In addition, Singh and co-workers have reported that when the protospacer is “preunwound” by introducing unpaired t-/nt-strand nucleotides (i.e., a DNA bubble), cleavage was prevented with RNA/DNA mismatches presented only at the two nucleotides immediately next to the PAM.32 This indicates that PAM-adjacent RNA/DNA mismatches prevent further unwinding of the protospacer to form the RNA/DNA hybrid even with pre-unwound (i.e., bubble) DNAs. In our work, the off-target bubble duplexes would not allow RNA/DNA hybrid formation, therefore their binding to Cas12a should not be attributed to “pre-unwinding” of the protospacer, but instead to flexibility induced by the bubble (Figure 4C).
In studies of CRISPR-Cas9, Severinov and co-workers showed that unpaired segments adjacent to PAM facilitate DNA deformation and significantly enhance the on-rate of binding.56 Rueda and co-workers reported direct observations of off-target bubble DNA binding, and correlated it with off-target events observed when the DNA is under mechanical distortion due to stretching with high force.53 DNA recognition by Cas12a22 and Cas946 share many similar mechanistic features, including PAM recognition, PAM-proximal distortion, as well as DNA unwinding and R-loop formation. This supports the conclusion from this work that Cas12a is also susceptible to promiscuous binding of flexible DNA duplexes.
While in vitro studies presented here demonstrate that PAM-adjacent protospacer flexibility leads to promiscuous DNA binding by Cas12a in the absence of RNA/DNA complementarity, it remains to be determined whether this feature is maintained in vivo. Genome-wide studies have shown that Cas9 binds to many more DNA sites in vivo than those that are cleaved.60−62 Coupled with the recent finding on Cas9 binding to flexible DNA,56 it has been proposed that duplex DNA destabilization during cellular processes, such as transcription and replication, may expose off-target sites to Cas9 activity.53 Given the similarity between Cas12a and Cas9, it is reasonable to hypothesize that variations of DNA flexibility between genomic sites or during different biological processes may influence in vivo Cas12a target discrimination, in particular, off-target DNA binding. As such, DNA flexibility may impact Cas12a-based applications, particularly those relying on Cas12a binding to specific DNA sites.16−20
In summary, data reported show that flexible DNA duplexes without target-strand/RNA-guide complementarity can bind to the site used by Cas12a for RNA-guided double-stranded DNA binding and cleavage. The results support the notion that flexibility and other physical properties of the DNA can influence target discrimination by CRISPR-Cas, including Cas12a, and therefore may impact developments of CRISPR-based technology.
Materials and Methods
Plasmids for Cas12a Protein Production
The plasmid used for LbCas12a expression encoded the full-length protein (residues 1–1274) with a fusion of an N-terminal 6× His tag followed by a maltose-binding protein (MBP) tag and a tobacco etch virus (TEV) cleavage site. The plasmid was a gift from Dr. F. Zhang6 (Addgene plasmid # 90096; http://n2t.net/addgene:90096; RRID: Addgene_90096). The plasmid used for FnCa12a expression encoded the full-length protein (residues 1–1300) with a fusion of N-terminal His8-3C protease-His7 tag followed by an MBP tag and a TEV cleavage site.33 The catalytically in-active dFnCas12a contained mutations of D917A and E1006A6 and was constructed using the FnCas12a plasmid with assistance from the Structural Biology Center at the Bridge Institute of USC. All plasmid sequences were confirmed prior to protein expression.
Cas12a Protein Expression and Purification
LbCas12a and FnCas12a were expressed in Escherichia coli. and purified following procedures similar to those previously reported.6,22,33 Briefly, T7 Express Competent E. coli (New England Biolabs, #C2566I) or Rosetta 2 (DE3) Competent E. coli. (Novagen) were transformed with the desired plasmids by heat shock. A single colony from the transformation was inoculated into Lysogeny Broth with an appropriate antibiotic (100 μg/mL ampicillin for LbCas12a and kanamycin for FnCas12a) and incubated at 37 °C overnight. Then, the small-scale culture was added to Terrific Broth with 100 μg/mL antibiotic (approximate 8 mL culture for a 1 L of cell growth) and incubated at 30 °C until the optical density at 600 nM (OD600) reached 0.5–0.6. Then the temperature was reduced to 18 °C, and incubation was continued for 20 min. Overexpression was then induced by adding 200 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) and shaking at 18 °C for 16–20 h.
The cells were harvested by centrifugation and re-suspended at 4 °C in lysis buffer [50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid, pH 7.0, 1 M NaCl, 5 mM MgCl2, 2 mM dithiothreitol (DTT), 10% glycerol] with protease inhibitor (Roche) and DNase. The cells were lysed at 4 °C, followed by ultracentrifugation at 4 °C to remove the cell debris. The supernatant was collected and was subjected to affinity chromatography (e.g., Ni-NTA). If desired, the MBP-His tag was cleaved with the TEV protease.33 Products from the first-round of affinity chromatography purification (and TEV cleavage if desired) were further purified by fast protein liquid chromatography using a size-exclusion S200 column (GE Healthcare). The collected fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and the fractions that contained the target protein was concentrated, exchanged into the storage buffer (20 mM Tris, pH 7.5, 200 mM KCl, 5 mM MgCl2, 0.5 mM TCEP, 10% glycerol), and stored at −80 °C. Also note that initial experiments were carried out with LbCas12a protein kindly provided by Dr. Ian M. Slaymaker (Broad Institute, Boston, MA), which has the MBP tag cleaved off. Experiments showed that the MBP tag does not influence the DNA cleavage activities investigated in this work (see Supporting Information Section S5).
AsCas12a protein was obtained commercially from Integrated DNA Technologies (Coralville, Iowa, #1081068).
Concentrations of proteins were determined according to absorbance at 280 nM with an extinction coefficient calculated from amino acids sequence: LbCas12a, 181 190 M–1·cm–1; LbCas12a with MBP tag, 249 030 M–1·cm–1; AsCas12a, 143 940 M–1·cm–1; and FnCas12a, 144 000 M–1·cm–1.
RNA Transcription
RNAs used in this study were synthesized by T7 in vitro transcription following a previously reported protocol.63 Sequences of the T7 top-strand primer and DNA templates used for transcribing different RNAs are listed in Table S2. A typical transcription reaction (400 μL) contained 0.5 μM DNA template, 1 μM T7 top-strand primer, 1 mM each of nucleotide tri-phosphate, 0.01% Triton, 400 units of T7 polymerase (e.g., New England Biolabs #M0251L), 40 mM tris(hydroxymethyl)aminomethane (Tris) pH 7.5, 15 mM MgCl2, 2 mM spermidine, and 5 mM DTT. The reaction mix was incubated at 37 °C for 3 h and then quenched by adding 20 mM ethylenediaminetetraacetic acid (EDTA). The RNA products were recovered by ethanol precipitation and then purified by denaturing PAGE. The final products were re-suspended in ME buffer [10 mM 3-(N-morpholino) propanesulfonic acid pH 6.5 and 1 mM EDTA] and stored at −20 °C.
Sequences of crRNA used in this study are listed in Table S1. The concentrations of RNA were determined according to absorbance at 260 nm. The molar extinction coefficients of RNA were estimated by ε = # of nucleotide × 10 000 M–1·cm–1.
Target DNA Constructs
Detailed design and sequences of the DNA substrate duplexes are described in Supporting Information Section S2. All DNA oligonucleotides were obtained by solid-phase chemical synthesis (Integrated DNA Technologies, Coralville, Iowa). To obtain a target DNA duplex, two desired strands were mixed in a 1:1 molar ratio in an annealing buffer (20 mM Tris pH 7.5, 100 mM KCl), the mixture was heated at 95 °C for 1 min, incubated at room temperature overnight, then stored at −20 °C until use.
DNA Cleavage Assay
To monitor DNA cleavage, one or both target strands were 32P labeled at the 5′ terminus following a previous protocol.64 A typical 10 μL 32P-labeling reaction contained 1 μM DNA strand, 1 μL 32P γ-ATP (MP Biomedicals, 6000 Ci/mmol), 10 units T4 polynucleotide kinase (PNK, New England Biolabs #M0201), and 1× PNK buffer (New England Biolabs, 70 mM Tris-HCl, 10 mM MgCl2, 5 mM DTT, pH 7.6). The reaction mixture was incubated at 37 °C for 30 min and then the T4 PNK was heat deactivated by incubation at 65 °C for 20 min. The excess 32P γ-ATP was not removed. Assuming no loss of 32P γ-ATP-labeled DNA, an equal amount of the unlabeled complementary strands was added to form a DNA duplex following the procedure described above.
In a typical single-turnover cleavage reaction, 32P-labeled DNA duplex (1–10 nM) were subjected to cleavage by a preformed Cas12a/crRNA effector complex, with the concentration of the complex at least 10 times higher than that of the DNA. To pre-form the effector complex, appropriate amount of the crRNA was first heated at 95 °C for 1 min and then incubated at room temperature in a reaction buffer (20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, 5% (v/v) glycerol, and 0.5 mM TCEP) for 10 min. The desired amount of Cas12a in the reaction buffer was added, so that the crRNA/Cas12a ratio was approximately 1.5:1. After incubating the Cas12a/crRNA mix at 37 °C for 15 min, an appropriate amount of DNA substrate was added, and the cleavage reaction was allowed to proceed at 37 °C for 30 min. Upon conclusion of the reaction, an equal volume of a denaturing dye solution (8 M urea, 20 mM EDTA, 20% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol) was added, and the mixture was immediately heated at 95 °C for 1 min to deactivate the enzyme.
The reaction products were resolved using denaturing PAGE, and DNA species were visualized and quantitated by autoradiography using a Personal Molecular Imager (Bio-Rad). The percentage of the product (% product) was calculated according to
| 1 |
where Iprecursor is the intensity of the DNA precursor signal and Iproduct is the product signal. Each particular signal I was corrected for background according to
| 2 |
where I0 is raw measured signal, S0 is the area of measured band, and abg is the average intensity per unit area obtained from multiple sections of the gel between the precursor and product bands.
Values of average and standard deviation were obtained from three or more replications.
Competition Assay
Competition experiments were typically carried out in the cleavage reaction buffer with 1 nM 32P-labeled probe duplex, 10 nM Cas12a/crRNA effector complex, and 1000 nM (1 μM) unlabeled competitor DNA duplex. To carry out a competition reaction, 32P-labeled probe duplex and unlabeled competitor DNA duplex were independently annealed as described above (i.e., “Target DNA Constructs”). The Cas12a/crRNA effector complex was preformed as described (i.e., “DNA Cleavage Assay”) and then added to a mixture containing the 32P-labeled probe and the unlabeled competitor. The reaction was allowed to proceed at 37 °C for 30 min and then terminated following procedures described (i.e., “DNA Cleavage Assay”). The DNA products were characterized and quantified as described above. Values of average and standard deviation were obtained from three or more replications.
Acknowledgments
We thank Dr. Ian Slaymaker (Broad Institute) for providing an initial protein sample and for suggestions on the manuscript, Dr. Lin Chen (USC) for advice, and the Structural Biology Center at the Bridge Institute of USC for assistance with protein mutagenesis, expression and purification.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01469.
Additional data on characterizing interactions of DNAs with Cas12a (PDF)
Accession Codes
UniProt protein ID: LbCas12a, A0A182DWE3; AsCas12a, U2UMQ6; FnCas12a, A0Q7Q2 NCBI protein ID: LbCas12a, WP_051666128.1; AsCas12a, WP_021736722.1; FnCas12a, WP_003040289.1.
Author Present Address
∥ GenapSys, Inc., 200 Cardinal Way, Redwood City, CA 94063.
The research was supported in part by grants from NIGMS (R01 GM124413, PZQ; P20GM103640, RR) and NSF (CHE-1213673, PZQ; MCB-1716423, RR).
The authors declare no competing financial interest.
Supplementary Material
References
- Koonin E. V.; Makarova K. S.; Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G. J.; Doudna J. A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M.; Chylinski K.; Fonfara I.; Hauer M.; Doudna J. A.; Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P.; Yang L.; Esvelt K. M.; Aach J.; Guell M.; DiCarlo J. E.; Norville J. E.; Church G. M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L.; Ran F. A.; Cox D.; Lin S.; Barretto R.; Habib N.; Hsu P. D.; Wu X.; Jiang W.; Marraffini L. A.; Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B.; Gootenberg J. S.; Abudayyeh O. O.; Slaymaker I. M.; Makarova K. S.; Essletzbichler P.; Volz S. E.; Joung J.; van der Oost J.; Regev A.; Koonin E. V.; Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S.; Larson M. H.; Gilbert L. A.; Doudna J. A.; Weissman J. S.; Arkin A. P.; Lim W. A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A.; Horlbeck M. A.; Adamson B.; Villalta J. E.; Chen Y.; Whitehead E. H.; Guimaraes C.; Panning B.; Ploegh H. L.; Bassik M. C.; Qi L. S.; Kampmann M.; Weissman J. S. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A. C.; Kim Y. B.; Packer M. S.; Zuris J. A.; Liu D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. B.; Komor A. C.; Levy J. M.; Packer M. S.; Zhao K. T.; Liu D. R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017, 35, 371–376. 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P.; Bryant E. E.; Joseph S. A.; Nambiar T. S.; Hayward S. B.; Rothstein R.; Ciccia A. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol. Cell 2017, 67, 1068–1079. 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Gilbert L. A.; Cimini B. A.; Schnitzbauer J.; Zhang W.; Li G.-W.; Park J.; Blackburn E. H.; Weissman J. S.; Qi L. S.; Huang B. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Kim J.; Hur J. K.; Been K. W.; Yoon S.-h.; Kim J.-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B. P.; Tsai S. Q.; Prew M. S.; Nguyen N. T.; Welch M. M.; Lopez J. M.; McCaw Z. R.; Aryee M. J.; Joung J. K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B.; Heidenreich M.; Mohanraju P.; Fedorova I.; Kneppers J.; DeGennaro E. M.; Winblad N.; Choudhury S. R.; Abudayyeh O. O.; Gootenberg J. S.; Wu W. Y.; Scott D. A.; Severinov K.; van der Oost J.; Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak Y. E.; Kleinstiver B. P.; Nuñez J. K.; Hsu J. Y.; Horng J. E.; Gong J.; Weissman J. S.; Joung J. K. Inducible and multiplex gene regulation using CRISPR–Cpf1-based transcription factors. Nat. Methods 2017, 14, 1163–1166. 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Lowder L. G.; Zhang T.; Malzahn A. A.; Zheng X.; Voytas D. F.; Zhong Z.; Chen Y.; Ren Q.; Li Q.; Kirkland E. R.; Zhang Y.; Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Han J.; Chen Z.; Wu H.; Dong H.; Nie G. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat. Commun. 2017, 8, 2095. 10.1038/s41467-017-02265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wang J.; Cheng Q.; Zheng X.; Zhao G.; Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discovery 2017, 3, 17018. 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Wang Y.; Liu Y.; Yang B.; Wang X.; Wei J.; Lu Z.; Zhang Y.; Wu J.; Huang X.; Yang L.; Chen J. Base editing with a Cpf1–cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- Dong D.; Ren K.; Qiu X.; Zheng J.; Guo M.; Guan X.; Liu H.; Li N.; Zhang B.; Yang D.; Ma C.; Wang S.; Wu D.; Ma Y.; Fan S.; Wang J.; Gao N.; Huang Z. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 2016, 532, 522–526. 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Yamano T.; Nishimasu H.; Zetsche B.; Hirano H.; Slaymaker I. M.; Li Y.; Fedorova I.; Nakane T.; Makarova K. S.; Koonin E. V.; Ishitani R.; Zhang F.; Nureki O. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P.; Yang H.; Rajashankar K. R.; Huang Z.; Patel D. J. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016, 26, 901–913. 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T.; Zetsche B.; Ishitani R.; Zhang F.; Nishimasu H.; Nureki O. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1. Mol. Cell 2017, 67, 633–645. 10.1016/j.molcel.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts D. C.; van der Oost J.; Jinek M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 2017, 66, 221–233. 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S.; Alcón P.; Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 2017, 546, 559–563. 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- Nishimasu H.; Yamano T.; Gao L.; Zhang F.; Ishitani R.; Nureki O. Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1. Mol. Cell 2017, 67, 139–147. 10.1016/j.molcel.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S.; Mesa P.; Thomsen J.; Paul B.; Alcón P.; Jensen S. B.; Saligram B.; Moses M. E.; Hatzakis N. S.; Montoya G. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 2018, 175, 1856–1871. 10.1016/j.cell.2018.10.045. [DOI] [PubMed] [Google Scholar]
- Swarts D. C.; Jinek M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell 2019, 73, 589–600. 10.1016/j.molcel.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohkendl I.; Saifuddin F. A.; Rybarski J. R.; Finkelstein I. J.; Russell R. Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a. Mol. Cell 2018, 71, 816–824. 10.1016/j.molcel.2018.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.; Choi Y. H.; Jang Y.; Yu J.; Goo J.; Lee G.; Jeong Y. K.; Lee S. H.; Kim I.-S.; Kim J.-S.; Jeong C.; Lee S.; Bae S. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018, 9, 2777. 10.1038/s41467-018-05245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.; Mallon J.; Poddar A.; Wang Y.; Tippana R.; Yang O.; Bailey S.; Ha T. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 5444–5449. 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan R.; Parameshwaran H. P.; Yogesha S. D.; Keilbarth M. W.; Rajan R. RNA-Independent DNA Cleavage Activities of Cas9 and Cas12a. Cell Rep. 2017, 21, 3728–3739. 10.1016/j.celrep.2017.11.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S.; Ma E.; Harrington L. B.; Da Costa M.; Tian X.; Palefsky J. M.; Doudna J. A., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–−439.. 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-Y.; Cheng Q.-X.; Liu J.-K.; Nie X.-Q.; Zhao G.-P.; Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Rep. 2018, 28, 491–493. 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. R.; Oakes B. L.; Sternberg S. H.; East-Seletsky A.; Kaplan M.; Doudna J. A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh O. O.; Gootenberg J. S.; Konermann S.; Joung J.; Slaymaker I. M.; Cox D. B. T.; Shmakov S.; Makarova K. S.; Semenova E.; Minakhin L.; Severinov K.; Regev A.; Lander E. S.; Koonin E. V.; Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A.; O’Connell M. R.; Knight S. C.; Burstein D.; Cate J. H. D.; Tjian R.; Doudna J. A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V.; Lin S.; Guilinger J. P.; Ma E.; Doudna J. A.; Liu D. R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D.; Scott D. A.; Weinstein J. A.; Ran F. A.; Konermann S.; Agarwala V.; Li Y.; Fine E. J.; Wu X.; Shalem O.; Cradick T. J.; Marraffini L. A.; Bao G.; Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.; Foden J. A.; Khayter C.; Maeder M. L.; Reyon D.; Joung J. K.; Sander J. D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Mao S.; Ying Y.; Krueger C. J.; Chen A. K. Progress and Challenges for Live-cell Imaging of Genomic Loci Using CRISPR-based Platforms. Genomics, Proteomics Bioinf. 2019, 17, 119–128. 10.1016/j.gpb.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P. G.; Barutcu A. R.; Shechner D. M.; Weiner C. L.; Melé M.; Rinn J. L. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat. Struct. Mol. Biol. 2018, 25, 176–184. 10.1038/s41594-017-0015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; Chen L.; Liu Y.-G. Off-target effects and the solution. Nat. Plants 2019, 5, 341–342. 10.1038/s41477-019-0406-z. [DOI] [PubMed] [Google Scholar]
- Tadić V.; Josipović G.; Zoldoš V.; Vojta A. CRISPR/Cas9-based epigenome editing: An overview of dCas9-based tools with special emphasis on off-target activity. Methods 2019, 164−165, 109–119. 10.1016/j.ymeth.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Jiang F.; Doudna J. A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Slaymaker I. M.; Gao L.; Zetsche B.; Scott D. A.; Yan W. X.; Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P.; Pattanayak V.; Prew M. S.; Tsai S. Q.; Nguyen N. T.; Zheng Z.; Joung J. K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S.; Dagdas Y. S.; Kleinstiver B. P.; Welch M. M.; Sousa A. A.; Harrington L. B.; Sternberg S. H.; Joung J. K.; Yildiz A.; Doudna J. A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 2017, 550, 407–410. 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P.; Sousa A. A.; Walton R. T.; Tak Y. E.; Hsu J. Y.; Clement K.; Welch M. M.; Horng J. E.; Malagon-Lopez J.; Scarfò I.; Maus M. V.; Pinello L.; Aryee M. J.; Joung J. K. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H.; LaFrance B.; Kaplan M.; Doudna J. A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 2015, 527, 110–113. 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu K.; Amrani N.; Jiang W.; Yogesha S. D.; Nguyen R.; Qin P. Z.; Rajan R. Bridge Helix of Cas9 Modulates Target DNA Cleavage and Mismatch Tolerance. Biochemistry 2019, 58, 1905–1917. 10.1021/acs.biochem.8b01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M. D.; Taylor B. J.; Driessen R. P. C.; Roos L.; Cvetesic N.; Allyjaun S.; Lenhard B.; Cuomo M. E.; Rueda D. S. DNA stretching induces Cas9 off-target activity. Nat. Struct. Mol. Biol. 2019, 26, 185–192. 10.1038/s41594-019-0188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; Jin X.; West S. M.; Joshi R.; Honig B.; Mann R. S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010, 79, 233–269. 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H.; Ran F. A.; Hsu P. D.; Konermann S.; Shehata S. I.; Dohmae N.; Ishitani R.; Zhang F.; Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V.; Minakhin L.; Severinov K. Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 5443–5448. 10.1073/pnas.1619926114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H.; Redding S.; Jinek M.; Greene E. C.; Doudna J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forties R. A.; Bundschuh R.; Poirier M. G. The flexibility of locally melted DNA. Nucleic Acids Res. 2009, 37, 4580–4586. 10.1093/nar/gkp442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Nguyen P.; Tangprasertchai N. S.; Reyes C. V.; Zhang X.; Qin P. Z., Nucleic acid structure and dynamics: perspectives from site-directed spin labeling. In Electron Paramagnetic Resonance; The Royal Society of Chemistry, 2015; Vol. 24, pp 122–147. [Google Scholar]
- Kuscu C.; Arslan S.; Singh R.; Thorpe J.; Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Wu X.; Scott D. A.; Kriz A. J.; Chiu A. C.; Hsu P. D.; Dadon D. B.; Cheng A. W.; Trevino A. E.; Konermann S.; Chen S.; Jaenisch R.; Zhang F.; Sharp P. A. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014, 32, 670–676. 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H.; Henry I. M.; Bhakta M. S.; Meckler J. F.; Segal D. J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015, 43, 3389–3404. 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangprasertchai N. S.; Di Felice R.; Zhang X.; Slaymaker I. M.; Vazquez Reyes C.; Jiang W.; Rohs R.; Qin P. Z. CRISPR–Cas9 Mediated DNA Unwinding Detected Using Site-Directed Spin Labeling. ACS Chem. Biol. 2017, 12, 1489–1493. 10.1021/acschembio.6b01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Cai Q.; Qin P. Z. The procapsid binding domain of phi29 packaging RNA has a modular architecture and requires 2’-hydroxyl groups in packaging RNA interaction. Biochemistry 2005, 44, 9348–9358. 10.1021/bi0475020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


