Abstract
Nanoparticle drug delivery applications have predominantly focused on the entrapment and delivery of hydrophobic molecules with poor water solubility. However, benefits can also be obtained from nanoparticle-based delivery of hydrophilic therapeutics. This study reports on the development of a p(HEMA-ran-GMA)-based nanoparticle synthesized via a spontaneous water-in-oil inverse nanoemulsion to deliver doxorubicin, a water-soluble chemotherapeutic. High drug loading efficiency and sustained release of doxorubicin from Cy5-functionalized p(HEMA-ran-GMA) nanoparticles enabled effective inhibition of the MCF-7 human breast cancer derived cell line. Direct comparative analyses with a hydrophobic PGMA nanoparticle demonstrated enhanced capabilities of the p(HEMA-ran-GMA)-based nanoparticle in vitro. The results suggest that p(HEMA-ran-GMA)-based nanoparticles, which are better suited for hydrophilic drug loading and delivery, may have the potential for the improved therapeutic effect in vivo by enhanced permeation and retention of the nanoparticles by avoidance of off-site side effects of the chemotherapeutic.
Introduction
The advent of nanomedicine is a promising alternative to overcome limitations of chemotherapy-mediated cancer therapy. Drug-loaded nanocarriers such as liposomes, polymer conjugates, polymer nanoparticles, and micelles1 can take advantage of the enhanced permeation and retention (EPR) effect due to the aberrant hypervascularization at tumorigenic sites, to increase the local concentration of the nanoencapsulated therapeutic cargo in tumors while reducing exposure of the drug to healthy cells.2 This tactic is proposed to increase the therapeutic index of chemotherapeutic agents such as doxorubicin (DOX) at the tumor site.
DOX, a potent, anthracycline antibiotic isolated from cultures of Streptomyces peucetius var. caesius,3 is frequently used as the first-line anti-tumor agent in the oncologic practice.4 Having the approval of the U.S. Food and Drug Administration (FDA) for its medical use, DOX has been indicated for administration as an intravenous bolus, either as a single agent or in combination with other chemotherapeutic agents for higher response rates and early onset of clinical benefits5 for the treatment of a variety of cancers such as acute myeloblastic leukemia, Hodgkin’s and non-Hodgkin’s lymphoma, esophageal carcinoma, osteosarcoma, and metastatic breast cancer.6 Liposomal formulations of DOX such as Doxil, Caelyx, and Myocet are currently in clinical use with evidence of lowered occurrences of DOX-related toxicity.7 Such nanoscaled lipid bilayer drug delivery vehicles have been extensively studied due to their capacity to effectively load and release both hydrophobic (e.g., paclitaxel) and hydrophilic (e.g., DOX) drugs.8,9 However, the introduction of liposomal drugs has been associated with palmar-plantar erythrodysesthesia (PPE) and mucositis, which can significantly impair the patient’s quality of life with no effective methods of prevention or treatment apart from dose reductions.10 Additionally, liposomal formulations have yet to address issues pertaining to the reversal of the multiple drug resistance (MDR) phenotype in tumor cells appropriately, which continues to be a hurdle in effective cancer treatment.1
Of the various nanoparticulate formulations being explored for therapeutic purposes, polymeric nanoparticles are highly attractive because of the versatility of the material, highly controllable physicochemical properties, relative ease, repeatability of production, and low cost. Good biocompatibility and biodegradability of polymers such as poly(lactic glycolic acid) (PLGA) and poly(caprolactone) (PCL) have also been indicated as an added advantage for therapeutic use as immune responses can be minimized.11 However, as hydrophilic drug delivery using polymeric nanoparticles can be challenging, largely due to the lack of interaction between the polymeric material and water-soluble drugs,12 and clinical translation of polymeric nanoparticulate systems to enhance treatment with hydrophilic drugs such as DOX has been hindered. This has been the case even with amphiphilic and well-established PLGA-based nanoparticles.11 The poor dispersion of hydrophilic drugs within the polymer matrix may result in inadequate drug loading and inefficient pharmacokinetics brought about by the “burst” effect when the polymeric nanoparticles are introduced into the physiological environment. The rapidly exposed drugs may also be prematurely subjected to hydrolytic degradation, leading to unsuitable biodistribution. Therefore, although polymer-based nanoparticles have had comparative success in the incorporation and delivery of hydrophobic drugs for therapy, there are still limitations in the development of formulations designed for the encapsulation and release of water-soluble drugs, creating a demand for the development of efficient hydrophilic nanoparticles for drug delivery. Surface wettability of nanoparticles is also a key factor to consider when developing therapeutic nanoparticles for cancer therapy, as hydrophobic nanoparticles have been known to preferentially accumulate various serum proteins on their surfaces, leading to opsonization by reticuloendothelial system (RES) recognition,13 which could potentially reduce the possibility of passive nanoparticle uptake at tumorigenic sites by the EPR effect.
Results and Discussion
In this study, hydrophilic polymeric nanoparticles were prepared using a water-soluble random copolymer, p(HEMA-ran-GMA), via a spontaneous water-in-oil (W/O) inverse nanoemulsion method (Scheme 1). The random copolymer was synthesized by atom transfer radical polymerization (ATRP) of 2-hydroxyethyl methacrylate (HEMA) and glycidyl methacrylate (GMA). Gel permeation chromatography (GPC) and 1H nuclear magnetic resonance (1H NMR) assessment of the copolymer indicated that the weighted average molecular weight was 29.3 kDa, and the copolymer backbone was consisted of ∼13% GMA (see Figure S1 in the Supporting Information). p(HEMA-ran-GMA) was deemed a suitable base material for the hydrophilic nanoparticle formulation due to the presence of hydroxyl functional groups from HEMA, which enables water-trapping capability, and the presence of highly reactive epoxide groups from GMA that may facilitate a wide range of nucleophilic ring-opening reactions for the functionalization.
Scheme 1. Synthesis of Cross-Linked p(HEMA-ran-GMA) Nanoparticles via Spontaneous Water-in-Oil (W/O) Inverse Nanoemulsion.

Nanoparticles were retrieved after formation by disruption of the emulsion through the addition of water.
As depicted in Scheme 1, the solubilization of p(HEMA-ran-GMA) in water was an important factor that supported the W/O spontaneous nanoemulsion method to produce cross-linked, hydrophilic polymeric nanoparticles. The use of the double-chained anionic surfactant, sodium dioctyl sulfosuccinate (AOT) as the surfactant, was advantageous due to its low water solubility and ability to stabilize the emulsion without the requirement of a further co-surfactant as used in other W/O systems.14 This is due to the fact that AOT, when dissolved in organic solvents, forms thermodynamically stable micelles consisting of a hydrophilic core that is compartmentalized by the hydrophilic head group of the AOT with the hydrophobic alkyl tails extending into the nonpolar continuous phase solvent.15 Ethylene diamine is a suitable cross-linking reagent to impart structural rigidity to the core of the nanoparticles in the emulsion, which are then readily retrieved as a suspension by centrifugation, after disrupting the stable W/O emulsion by the addition of excess Milli-Q water. Hydrophobic poly(glycidyl methacrylate) (PGMA)-based nanoparticles were assessed in parallel in this study. PGMA nanoparticles were synthesized by the solvent evaporation oil-in-water (O/W) emulsion method, and the justifications for their use for therapeutic purposes have been previously described extensively.16−18
Both p(HEMA-ran-GMA)- and PGMA-based nanoparticles synthesized for this study were functionalized with a Cyanine5 (Cy5) fluorophore to allow detection and tracking by fluorescent confocal imaging (see Schematic S1 in the Supporting Information). Table 1 summarizes the data obtained for the nanoparticle variants’ hydrodynamic sizes using dynamic light scattering (DLS) and the respective surface charges. Figure S2 in the Supporting Information shows the nanoparticle variants’ size distribution with respect to scattering light intensity. It could be observed that the surface charge of the p(HEMA-ran-GMA) nanoparticle became less negative as the functionalization proceeded stepwise toward the Cy5 conjugation, indicating changes in surface modification of the nanoparticle at each stage. The overall negative surface charges of the p(HEMA-ran-GMA)-based nanoparticles in comparison to the positively charged Cy5-PGMA nanoparticles (+39 mV) may be explained by the presence of hydroxyl groups (−OH) on the nanoparticle surface that dissociate readily in an aqueous medium.
Table 1. Physicochemical Properties of PGMA- and p(HEMA-ran-GMA)-Based Nanoparticles Assessed Using Dynamic Light Scattering (DLS)a.
| nanoparticle type | mean hydrodynamic diameter (d.nm) (PDI) | ζ potential (mV) (SD) |
|---|---|---|
| Cy5-PGMA | 166 (0.109) | +39 (3.59) |
| p(HEMA-ran-GMA)b | 271 (0.198) | –75 (15.5) |
| NH2-functionalized p(HEMA-ran-GMA)c | 230 (0.105) | –48 (20.5) |
| Cy5-p(HEMA-ran-GMA) | 244 (0.137) | –12.5 (7.86) |
The table summarizes the mean hydrodynamic size, nanoparticle polydispersity index (PDI), and surface charge by means of ζ potential measurements (with standard deviation (SD)) of the nanoparticle suspensions in 1× phosphate-buffered saline (PBS) at pH 7.4 (N = 3).
Cross-linked p(HEMA-ran-GMA) nanoparticle as retrieved from water-in-oil (W/O) nanoemulsion.
Intermediate, amine-functionalized cross-linked p(HEMA-ran-GMA) nanoparticle before conjugation with Cy5 fluorophore via N-hydroxysuccinimide (NHS) ester.
Although all the nanoparticle variants stated in Table 1 were highly monodispersed as discerned from the polydispersity indexes (PDI) values, it was apparent that the hydrophilic p(HEMA-ran-GMA)-based nanoparticles were larger in size than the Cy5-PGMA nanoparticles. Transmission electron microscopy (TEM) images of both Cy5-PGMA and Cy5-p(HEMA-ran-GMA) nanoparticles are also provided in Figure 1A,B, respectively, to show the size differences. The augmented sizes of p(HEMA-ran-GMA)-based nanoparticles could be attributed to the hydrophilic material capacity to swell when hydrated unlike the hydrophobic PGMA-based nanoparticles. Additionally, it should be noted that the p(HEMA-ran-GMA) chains within the aqueous micelles in the W/O emulsion were cross-linked based on theoretical calculations that involved a coupling of ∼50 mol % of epoxide groups. It may be possible that increasing the cross-linking density within the polymer chains could have led to changes in a nanoparticle size; however, this was not attempted for this study. Previous studies have demonstrated that the size of the nanoparticles synthesized from W/O emulsions may be controlled by factors such as the water content in the aqueous phase, type of solvent used in the organic phase, choice of a surfactant (or a co-surfactant), and concentrations of the reagents used.19 In AOT-stabilized W/O spontaneous inverse nanoemulsions, it has been also suggested that the Wo ratio (Wo = [water]/[AOT] in an organic solvent) could play a role in controlling the nanoparticle size. No significant changes in the hydrodynamic size of the unfunctionalized and cross-linked p(HEMA-ran-GMA) nanoparticles were detected when the Wo ratio was varied from 0.5 to 10 while keeping the cross-linking density consistent with the addition of 50 mol % ethylene diamine (see Figure S3 in the Supporting Information). This lack of size variability with changing Wo suggests that this system may be more dependent on the combined effects of the solvent, surfactant, and polymer concentration in the aqueous phase as the driving factors that govern a nanoparticle size from the emulsion. As a result, it was determined that Wo = 10 was suitable for this nanoparticle synthesis system based on the economy of the reagent used and yield of nanoparticles achieved per batch.
Figure 1.
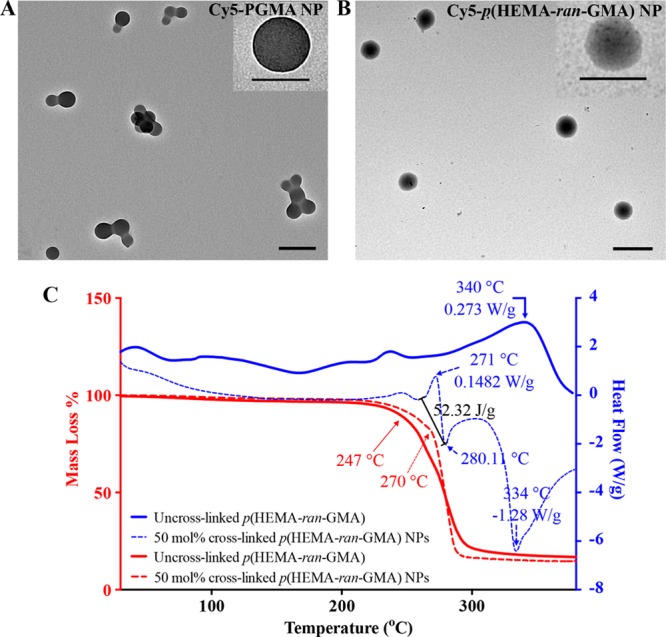
(A) Transmission electron microscopy (TEM) image of Cy5-PGMA nanoparticles (NPs). Scale bar = 200 nm. Inset: TEM image (top right), scale bar = 120 nm. (B) TEM image of p(HEMA-ran-GMA) nanoparticles. Scale bar = 500 nm. Inset: TEM image (top right), scale bar = 300 nm. (C) Thermal analysis of cross-linked p(HEMA-ran-GMA) nanoparticles with respect to p(HEMA-ran-GMA) polymer. Mass loss (%) was determined using thermogravimetric analysis (TGA) (in red), and heat flow assessment was performed using differential scanning calorimetry (DSC) (in blue) over a temperature range of 25 to 380 °C under inert conditions.
The thermal analysis of a polymeric material extracted from the W/O emulsions with and without the addition of ethylene diamine was conducted using thermogravimetric analysis (TGA) and differential scanning calorimetry (DSC) (Figure 1C) to assess the cross-linking capacity of ethylene diamine. From the TGA plots (in red), a delayed inflection point of the decomposition at 270 °C in the sample prepared with the addition of ethylene diamine suggests successful cross-linking. Furthermore, at approximately the same temperature, this material exhibited an endothermic reaction (+52.32 J/g at 271 °C) as observed by the DSC plots (in blue) compared to the sample prepared without ethylene diamine. Taken together, it could be inferred that ethylene diamine effectively assisted in coupling epoxide functional groups on the polymer backbone, resulting in discrete cross-linked p(HEMA-ran-GMA) nanoparticles within the AOT-stabilized micelles in the W/O emulsion.
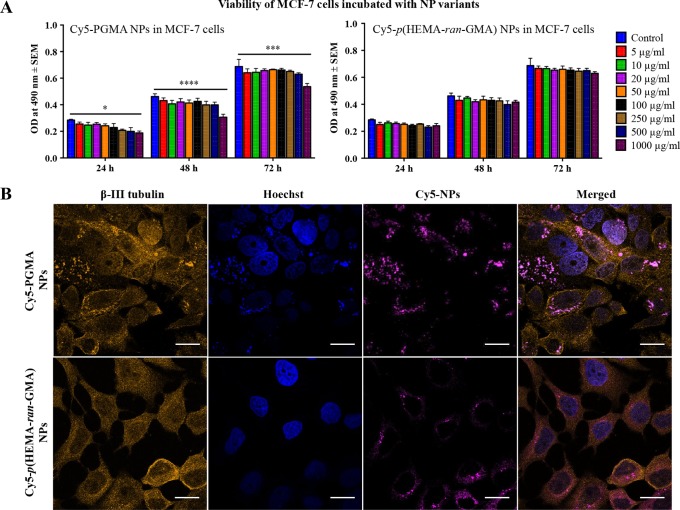
Cytotoxicity of Cy5-p(HEMA-ran-GMA) and Cy5-PGMA nanoparticles in a biologically relevant human breast cancer model using MCF-7 cells was assessed in vitro using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay over a course of 72 h, using various nanoparticle concentrations up to 1 mg/mL. All nanoparticle formulations were nontoxic across concentrations tested except for the Cy5-PGMA nanoparticles at the highest concentration of 1 mg/mL (Figure 2A). Confocal imaging of MCF-7 cells incubated with Cy5-functionalized nanoparticles showed that the cells sequestered both nanoparticle types (Figure 2B). The literature has stated that nanoparticle surface characteristics such as surface charge and hydrophobicity may be key requisites for cellular uptake.20 With cell membranes being negatively charged, it is widely accepted that the probability for positively charged nanoparticles to be internalized by cells is much higher. In tandem, confocal images of the MCF-7 cells incubated with equal concentration (20 μg/mL) of nanoparticle variants appeared to indicate preferential accumulation of the Cy5-PGMA nanoparticles within the cell body in comparison to Cy5-p(HEMA-ran-GMA) nanoparticles.
Figure 2.
In vitro assessment of Cy5-conjugated PGMA and p(HEMA-ran-GMA) nanoparticles (NPs) in MCF-7 cells. (A) Mean ± standard error of the mean (SEM) cytotoxicity of varying concentrations (0–1000 μg/mL) of Cy5-conjugated nanoparticles measured by the availability of viable MCF-7 cells ascertained by absorbance (OD: optical density) at 490 nm by MTS assay at 24, 48, and 72 h. Statistical analysis by two-way ANOVA with post hoc analysis using Dunnett’s multiple comparison test (n = 3; *p < 0.05, ***p < 0.0001, ****p < 0.00001). (B) Confocal images of MCF-7 cells incubated with 20 μg/mL of Cy5-conjugated NPs overnight. Z-stack images were obtained, and images are of single optical slices focused on the nuclear membrane (scale bars = 20 μm).
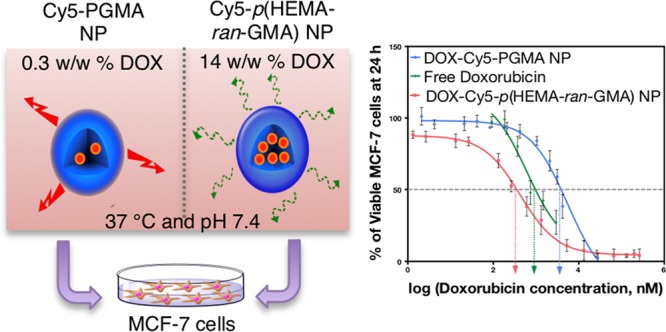
DOX was loaded into both Cy5-conjugated nanoparticle variants used in this study by a backfilling method that has been detailed in Section 2.6 of the Supporting Information. This technique was chosen to keep the drug loading method consistent between both Cy5-conjugated p(HEMA-ran-GMA) and PGMA nanoparticles that were synthesized by two different emulsion techniques. The backfilling of DOX was also an attractive option as it limits the exposure of the drug to reaction conditions necessary for nanoparticle synthesis, which could potentially affect drug activity (i.e., alkaline pH and elevated temperature).21 High-performance liquid chromatography (HPLC) was used to assess the loading efficiency of DOX and the drug release profiles from the nanoparticle variants (Figure 3). It was observed that the loading of DOX in Cy5-p(HEMA-ran-GMA) nanoparticles (∼14% w/w) was significantly higher than that in Cy5-PGMA nanoparticles (∼0.3% w/w). The higher drug loading efficiency in Cy5-p(HEMA-ran-GMA) nanoparticles may be explained by the enhanced interaction of the water-soluble drug with the hydrophilic nature of the HEMA groups in the base material, p(HEMA-ran-GMA). This explanation was further supported by the assessment of the release of DOX from both the nanoparticle variants. DOX-loaded Cy5-PGMA nanoparticles depicted a “burst” effect that almost instantaneously released more than 50% of loaded DOX, suggesting that the aqueous DOX interacted poorly with the hydrophobic core of the Cy5-PGMA nanoparticle, preferentially releasing the therapeutic contents into the exposed aqueous environment. The release profile of DOX from Cy5-p(HEMA-ran-GMA) nanoparticles was more sustained and gradual in comparison (over several days), suggesting that drug release from the DOX reservoir within the hydrophilic nanoparticle core could be dependent solely on diffusion by a concentration gradient with respect to the volume of the external aqueous sink (Figure 3). Correspondingly, a relevant study revealed that DOX release from p(HEMA) nanoparticles was controlled by the swelling capacity of the hydrophilic polymeric material, which varied with pH.22 Although it was critical for various studies based on pH-responsive drug delivery vehicles to report the effects of pH on DOX release,23−26 drug loading and release assessments conducted for these backfilled nanoparticle variants designed for passive accumulation at tumorigenic sites were limited to a normal physiological pH of 7.4.
Figure 3.
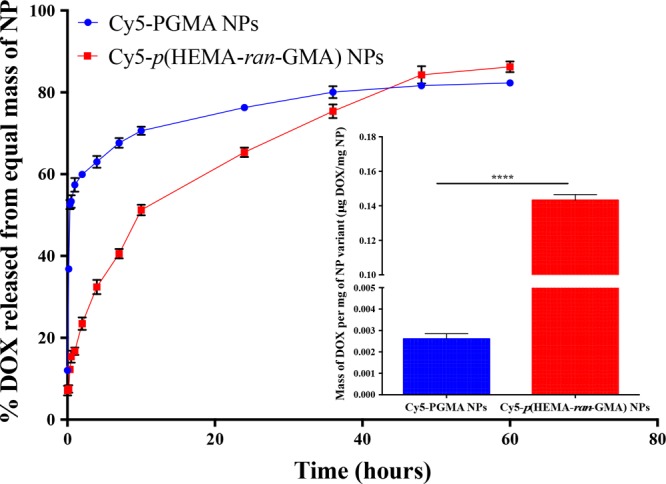
Doxorubicin (DOX) release profiles from Cy5-PGMA and Cy5-p(HEMA-ran-GMA) nanoparticles (NPs) assessed using high-performance liquid chromatography (HPLC) (sink volume = 10 mL; 200 μL sample assessed at each time point). Inset graph: DOX loading efficiency (DOX mg/nanoparticle mg ± standard error of measurement (SEM)). Statistical analysis by unpaired t test; ****p < 0.0001. n = 3 for both DOX loading and release profile measurements.
The therapeutic efficiency of the DOX-loaded nanoparticle variants compared to free drug was assessed in vitro to determine their half-maximal inhibitory concentration (IC50) in the MCF-7 cell line (Figure 4). The IC50 is a fundamental quantitative measure in pharmacology to indicate the potency of a drug in inhibiting a specific biological or biochemical function.27 After 24 h incubation of the DOX-loaded nanoparticles in MCF-7 cells, it was revealed that DOX-Cy5-PGMA nanoparticles had a significant higher IC50 than both free DOX and DOX-Cy5-p(HEMA-ran-GMA) nanoparticles. Although the IC50 of DOX-Cy5-p(HEMA-ran-GMA) nanoparticles was lower than that of free DOX, the difference was not statistically significant (see Figure S4 in the Supporting Information). Despite the outcome from this short term in vitro experiment, it is anticipated that the p(HEMA-ran-GMA)-based nanoparticle could have added advantages in vivo due to the EPR effect as well as the sustained release of the DOX from the nanoparticles over several days. (Figure 3: DOX release data represented with respect to time in hours). To obtain the desired 50% inhibitory effect by DOX-Cy5-PGMA nanoparticles, it was extrapolated from the DOX loading data (refer to inset graph in Figure 3) that an amount above the toxic threshold of the nanoparticle was required (1131.33 μg/mL). Therefore, the use of DOX-Cy5-PGMA nanoparticles developed in this study would not be recommended for therapeutic use as nanoparticle-associated cytotoxicity could override any inhibitory effect of DOX. As such, the IC50 determined from DOX-Cy5-PGMA nanoparticles may not be an accurate representation. In comparison, a therapeutic effect could be observed with a substantially smaller concentration (1.63 μg/mL) of DOX-loaded p(HEMA-ran-GMA) nanoparticles. These data suggest that the biocompatible and hydrophilic p(HEMA-ran-GMA)-based nanoparticles can be highly effective in encapsulating and delivering water-soluble chemotherapeutics such as DOX for noninvasive treatment.
Figure 4.
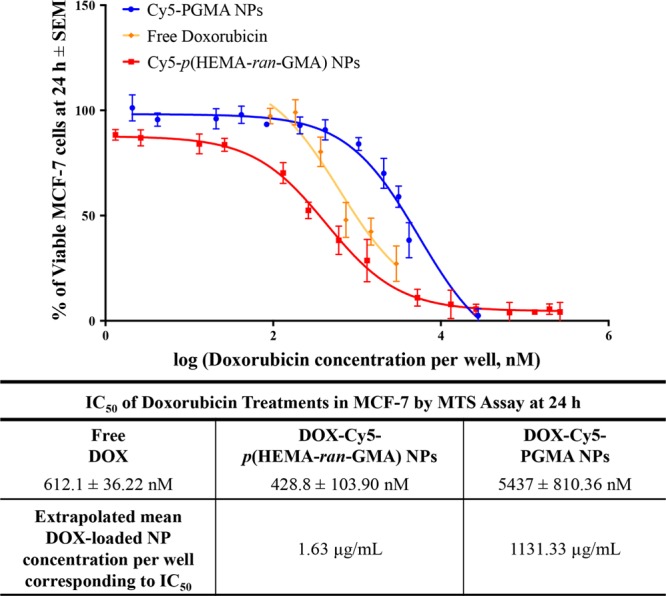
(Top panel) DOX (free and nanoparticle (NP)) dose–response regression curve of mean viability of MCF-7 cells at 24 h ± SEM (n = 3). (Bottom panel) Summary of mean IC50 values (± SEM) of DOX treatments used in the study and extrapolated concentrations of DOX-loaded nanoparticles correlating to IC50 values.
Conclusions
In conclusion, this study demonstrates the development of a hydrophilic polymer nanoparticle synthesized using a water-soluble copolymer, p(HEMA-ran-GMA), employing a W/O spontaneous inverse nanoemulsion. These hydrophilic nanoparticles are biocompatible at therapeutically relevant concentrations with the capacity for high drug loading of the water-soluble chemotherapeutic, DOX. The hydrophilicity of the nanoparticles coupled with sustained drug release could potentially enable prolonged circulation in systemic conditions such that uptake at tumorigenic sites via the EPR effect may be possible. The study has also confirmed the incompatibility of utilizing a hydrophobic polymeric nanoparticle such as the PGMA-based nanoparticle for the loading and delivery of water-soluble therapeutic agents.
Experimental Section
p(HEMA-ran-GMA) Random Copolymer Synthesis and Characterization
HEMA and GMA monomers were used in the ATRP synthesis of p(HEMA-ran-GMA). The random copolymerization reaction was carried out under Schlenk conditions at 80 °C for 2 h, with the addition of copper(I) bromide and 2,2-bypyridine. (4-Morpholino)-ethyl-2-bromoisobutyrate was added as an initiator. Purified p(HEMA-ran-GMA) was characterized by 1H nuclear magnetic resonance (NMR) and gel permeation chromatography (GPC).
Nanoparticle Synthesis and Characterization
PGMA Nanoparticle Synthesis
One hundred milligrams of PGMA was dissolved in the 1:3 mixture of chloroform and methyl ethyl ketone to form 8 mL of the organic phase. This was added dropwise into the aqueous phase with vigorous stirring made up of 30 mL of 1.25% w/v Pluronic F-108 in Milli-Q water and sonicated extensively. An aqueous suspension of PGMA nanoparticles was retrieved by removing all solvents under the reduced pressure at 40 °C.
p(HEMA-ran-GMA) Nanoparticle Synthesis
One hundred milligrams of p(HEMA-ran-GMA) (100 mg) was dissolved in 4 mL of Milli-Q water and added to a mixture of 17 g of sodium dioctyl sulfosuccinate in 250 mL of dry hexane to obtain an optically clear and homogeneous emulsion with moderate stirring. Forty-two microliters of 1:100 ethylene diamine was added to the emulsion and allowed to react overnight at room temperature. The cross-linked p(HEMA-ran-GMA) nanoparticles were retrieved by disrupting the emulsion with the addition of excess Milli-Q water and centrifugation. p(HEMA-ran-GMA) nanoparticles were purified by dialysis against Milli-Q water overnight.
Cy5 Functionalization of Nanoparticles
Both PGMA and p(HEMA-ran-GMA) nanoparticles were subjected to amine functionalization with excess aqueous ammonia before the Cy5 functionalization using Cy5-NHS ester. Fluorescent Cy5-conjugated nanoparticles were purified by dialysis against Milli-Q water.
Nanoparticle Characterization
Synthesized nanoparticles were characterized using dynamic light scattering, fluorescence measurements, and transmission light microscopy. Cy5-conjugated, cross-linked p(HEMA-ran-GMA) nanoparticles were additionally assessed using thermogravimetric analysis and differential scanning calorimetry.
Doxorubicin Loading and Release Assessments
Nanoparticle variants were backfilled with doxorubicin according to detailed procedures outlined in the Supporting Information. Drug loading and release profiles at physiologically relevant conditions (37 °C, pH 7.4) were assessed using high-performance liquid chromatography coupled with a UV/vis detector by an isocratic solvent system consisting of 0.02 M phosphate buffer (pH 5.4) and acetonitrile at a flow rate of 10 mL/min, to detect the doxorubicin peak at a retention time of 2 min at 233 nm.
Culture of MCF-7 Cells
MCF-7 cells (human breast adenocarcinoma cell line, ATCC) were cultured in minimum essential media α (MEM α, Gibco) supplemented with 0.15% sodium bicarbonate, 10% fetal bovine serum (FBS), and 1× GlutaMAX and were grown in a humidified incubator at 37 °C with 5% CO2.
Cytotoxicity Assessment
Cytotoxicity assessments and cell growth inhibition assessments were carried out on cultured MCF-7 cells (106 cells/well) in 96-well plates by the MTS assay using the protocol as described by the manufacturer (Promega CellTiter 96 AQueous One Solution Cell Proliferation Assay). The absorbance of the colored formazan product generated by viable cells was assessed on a plate reader at 490 nm.
Confocal Imaging
MCF-7 cells were seeded (5 × 104 cells/well) on poly(l-lysine)-treated cover slips (10 mm) in 24 well plates with 500 μL of culture media per well and allowed to settle overnight. Cy5-conjugated nanoparticle variants (20 μg/mL in 100 μL of culture media) were added in duplicate to the respective wells and incubated for 24 h. To fix the cells in each well, media were removed, cell layers were washed with 1× PBS twice, and then 4% w/v paraformaldehyde in 1× PBS was added for 15 min. After removing the fixative, the cover slips in each well were washed twice with 1× PBS before permeabilizing the cells in each well with 0.5% v/v Triton X-100 in 1× PBS for 15 min at room temperature. The permeabilized solution was removed, and each well was washed twice with 1× PBS before the addition of the 2% v/v donkey serum in 0.5% Triton X-100 solution (2% DKS) to block the cells for 1 h at 4 °C. Antibodies recognizing mouse βIII-tubulin (1: 500) in 2% DKS were added to each well and incubated overnight at 4 °C. After removing the primary antibodies, Hoechst stain (1:2000) and AF555 goat anti-mouse secondary antibody (1:1000) in 2% DKS were added to each well and incubated in the dark at room temperature for 20 min. Prepared samples on cover slips were washed twice with 1× PBS and mounted onto glass slides for confocal imaging using spectral properties of DAPI (cell nucleus), AF555 (microtubules in cytoskeleton), and Cy5 (nanoparticles).
Statistical Analysis
All statistical analyses were conducted using GraphPad Prism 6.0.
Acknowledgments
The authors would like to acknowledge Mr. Pengfei Liu, Dr. Ruhani Singh, Mrs. Carole Bartlett, Mr. Guy Ben-Ary, Miss Sushmita Raja, Miss Jessica Kretzmann, Dr. Cameron Evans, Dr. Nicole M. Smith, and Professor Martin Saunders for their technical expertise and discussions.
Glossary
Abbreviations
- EPR
enhanced permeation and retention
- DOX
doxorubicin
- PGMA
poly(glycidyl methacrylate)
- p(HEMA-ran-GMA)
poly(hydroxyethyl methacrylate-glycidyl methacrylate)
- W/O
water-in-oil
- AOT
sodium dioctyl sulfosuccinate
- Cy5
Cyanine5
- PBS
phosphate-buffered saline
- DLS
dynamic light scattering
- TEM
transmission electron microscopy
- TGA
thermogravimetric Analysis
- DSC
differential scanning calorimetry
- HPLC
high-performance liquid chromatography
- GPC
gel permeation chromatography
- NMR
nuclear magnetic resonance
- NP
nanoparticle
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b02894.
Detailed experimental procedures and supporting figures (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
Australian Research Council and NHMRC Career Development Fellowship APP1087114.
The authors declare no competing financial interest.
Supplementary Material
References
- Ma P.; Mumper R. J. Anthracycline nano-delivery systems to overcome multiple drug resistance: A comprehensive review. Nano Today 2013, 8, 313–331. 10.1016/j.nantod.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greish K. Enhanced permeability and retention of macromolecular drugs in solid tumors: A royal gate for targeted anticancer nanomedicines. J. Drug Targeting 2007, 15, 457–464. 10.1080/10611860701539584. [DOI] [PubMed] [Google Scholar]
- Arcamone F.; Cassinelli G.; Fantini G.; Grein A.; Orezzi P.; Pol C.; Spalla C. Adriamycin, 14-hydroxydaimomycin, a new antitumor antibiotic from S. Peucetius var. caesius. Biotechnol. Bioeng. 1969, 11, 1101–1110. 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- Minotti G.; Menna P.; Salvatorelli E.; Cairo G.; Gianni L. Anthracyclines: Molecular Advances and Pharmacologic Developments in Antitumor Activity and Cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- Pagani O.; Senkus E.; Wood W.; Colleoni M.; Cufer T.; Kyriakides S.; Costa A.; Winer E. P.; Cardoso F. International guidelines for management of metastatic breast cancer: can metastatic breast cancer be cured?. J. Natl. Cancer Inst. 2010, 102, 456–463. 10.1093/jnci/djq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal P. K.; Iliskovic N. Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 1998, 339, 900–905. 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- Vail D. M.; Amantea M. A.; Colbern G. T.; Martin F. J.; Hilger R. A.; Working P. K. Pegylated liposomal doxorubicin: proof of principle using preclinical animal models and pharmacokinetic studies. Semin. Oncol. 2004, 16–35. 10.1053/j.seminoncol.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Koudelka Š.; Turánek J. Liposomal paclitaxel formulations. J. Controlled Release 2012, 163, 322–334. 10.1016/j.jconrel.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Niu G.; Cogburn B.; Hughes J.. Preparation and characterization of doxorubicin liposomes. In Cancer Nanotechnology; Springer: 2010; pp 211–219. [DOI] [PubMed] [Google Scholar]
- Shafei A.; El-Bakly W.; Sobhy A.; Wagdy O.; Reda A.; Aboelenin O.; Marzouk A.; El Habak K.; Mostafa R.; Ali M. A.; Ellithy M. A review on the efficacy and toxicity of different doxorubicin nanoparticles for targeted therapy in metastatic breast cancer. Biomed. Pharmacother. 2017, 95, 1209–1218. 10.1016/j.biopha.2017.09.059. [DOI] [PubMed] [Google Scholar]
- Vrignaud S.; Benoit J.-P.; Saulnier P. Strategies for the nanoencapsulation of hydrophilic molecules in polymer-based nanoparticles. Biomaterials 2011, 32, 8593–8604. 10.1016/j.biomaterials.2011.07.057. [DOI] [PubMed] [Google Scholar]
- Barichello J. M.; Morishita M.; Takayama K.; Nagai T. Encapsulation of Hydrophilic and Lipophilic Drugs in PLGA Nanoparticles by the Nanoprecipitation Method. Drug Dev. Ind. Pharm. 1999, 25, 471–476. 10.1081/DDC-100102197. [DOI] [PubMed] [Google Scholar]
- Duan X.; Li Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 2013, 9, 1521–1532. 10.1002/smll.201201390. [DOI] [PubMed] [Google Scholar]
- Banerjee T.; Mitra S.; Singh A. K.; Sharma R. K.; Maitra A. Preparation, characterization and biodistribution of ultrafine chitosan nanoparticles. Int. J. Pharm. 2002, 243, 93–105. 10.1016/S0378-5173(02)00267-3. [DOI] [PubMed] [Google Scholar]
- Cason J. P.; Miller M. E.; Thompson J. B.; Roberts C. B. Solvent effects on copper nanoparticle growth behavior in AOT reverse micelle systems. J. Phys. Chem. B 2001, 105, 2297–2302. 10.1021/jp002127g. [DOI] [Google Scholar]
- Lozić I.; Hartz R. V.; Bartlett C. A.; Shaw J. A.; Archer M.; Naidu P. S. R.; Smith N. M.; Dunlop S. A.; Iyer K. S.; Kilburn M. R.; Fitzgerald M. Enabling dual cellular destinations of polymeric nanoparticles for treatment following partial injury to the central nervous system. Biomaterials 2016, 74, 200–216. 10.1016/j.biomaterials.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Naidu P. S. R.; Norret M.; Smith N. M.; Dunlop S. A.; Taylor N. L.; Fitzgerald M.; Iyer K. S. The Protein Corona of PEGylated PGMA-Based Nanoparticles is Preferentially Enriched with Specific Serum Proteins of Varied Biological Function. Langmuir 2017, 33, 12926–12933. 10.1021/acs.langmuir.7b02568. [DOI] [PubMed] [Google Scholar]
- Evans C. W.; Viola H. M.; Ho D.; Hool L. C.; Dunlop S. A.; Fitzgerald M.; Iyer K. S. Nanoparticle-mediated internalisation and release of a calcium channel blocker. RSC Adv. 2012, 2, 8587–8590. 10.1039/c2ra21058d. [DOI] [Google Scholar]
- Eastoe J.; Hollamby M. J.; Hudson L. Recent advances in nanoparticle synthesis with reversed micelles. Adv. Colloid Interface Sci. 2006, 128-130, 5–15. 10.1016/j.cis.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577. 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen M. J. H.; Crommelin D. J. A.; Storm G.; Hulshoff A. Doxorubicin decomposition on storage. Effect of pH, type of buffer and liposome encapsulation. Int. J. Pharm. 1985, 23, 1–11. 10.1016/0378-5173(85)90217-0. [DOI] [Google Scholar]
- Chouhan R.; Bajpai A. K. Real time in vitro studies of doxorubicin release from PHEMA nanoparticles. J. Nanobiotechnol. 2009, 7, 5. 10.1186/1477-3155-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R.; Meng F.; Deng C.; Klok H.-A.; Zhong Z. Dual and multi-stimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials 2013, 34, 3647–3657. 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- Du C.; Deng D.; Shan L.; Wan S.; Cao J.; Tian J.; Achilefu S.; Gu Y. A pH-sensitive doxorubicin prodrug based on folate-conjugated BSA for tumor-targeted drug delivery. Biomaterials 2013, 34, 3087–3097. 10.1016/j.biomaterials.2013.01.041. [DOI] [PubMed] [Google Scholar]
- Du J.-Z.; Du X.-J.; Mao C.-Q.; Wang J. Tailor-Made Dual pH-Sensitive Polymer–Doxorubicin Nanoparticles for Efficient Anticancer Drug Delivery. J. Am. Chem. Soc. 2011, 133, 17560–17563. 10.1021/ja207150n. [DOI] [PubMed] [Google Scholar]
- Muhammad F.; Guo M.; Qi W.; Sun F.; Wang A.; Guo Y.; Zhu G. pH-triggered controlled drug release from mesoporous silica nanoparticles via intracelluar dissolution of ZnO nanolids. J. Am. Chem. Soc. 2011, 133, 8778–8781. 10.1021/ja200328s. [DOI] [PubMed] [Google Scholar]
- Sebaugh J. L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. 10.1002/pst.426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.