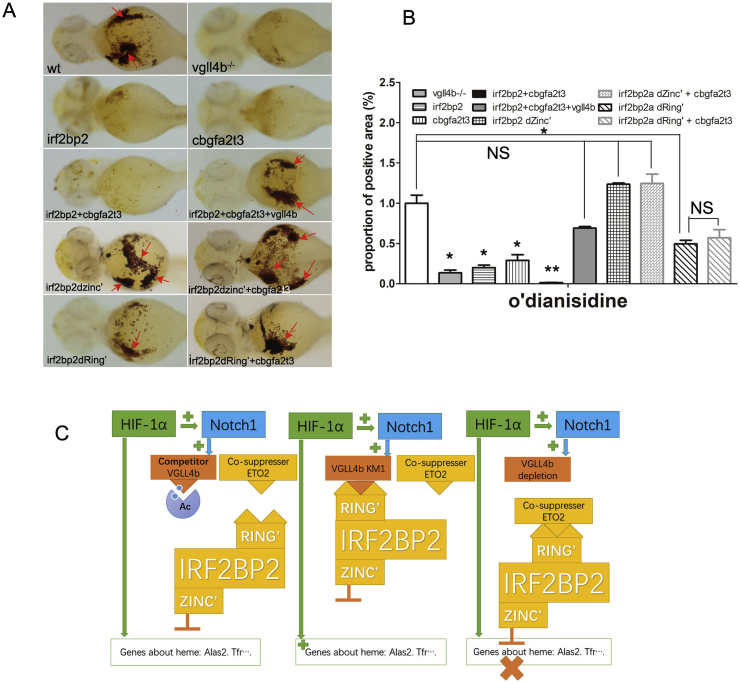
Fig. 7.
Bioeffects of IRF2BP2 mutants in synergistic effect with ETO2. (A) Bioeffects of IRF2BP2a mutants on heme biosynthesis suppression in addition to ETO2 detected by o’dianisidine stain. (B) Staining positive area compare among subgroups of IRF2BP2 mutants, depletion of either Zinc Finger or Ring Finger can abolish suppress effect of ETO2. (C) Model for the regulation of heme biosynthesis by alas2 et al., regulated by HIF1α, VGLL4 and IRF2BP2. HIF1α induced alas2 expression dependent on NOTCH1 and VGLL4. VGLL4 induced alas2 expression by reducing the IRF2BP2 transcription suppression and uncoupling the co-suppressor effects of IRF2BP2 and its partner ETO2. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *p<0. 05, **p<0.01, ***p<0.001. NS denotes no significant difference.