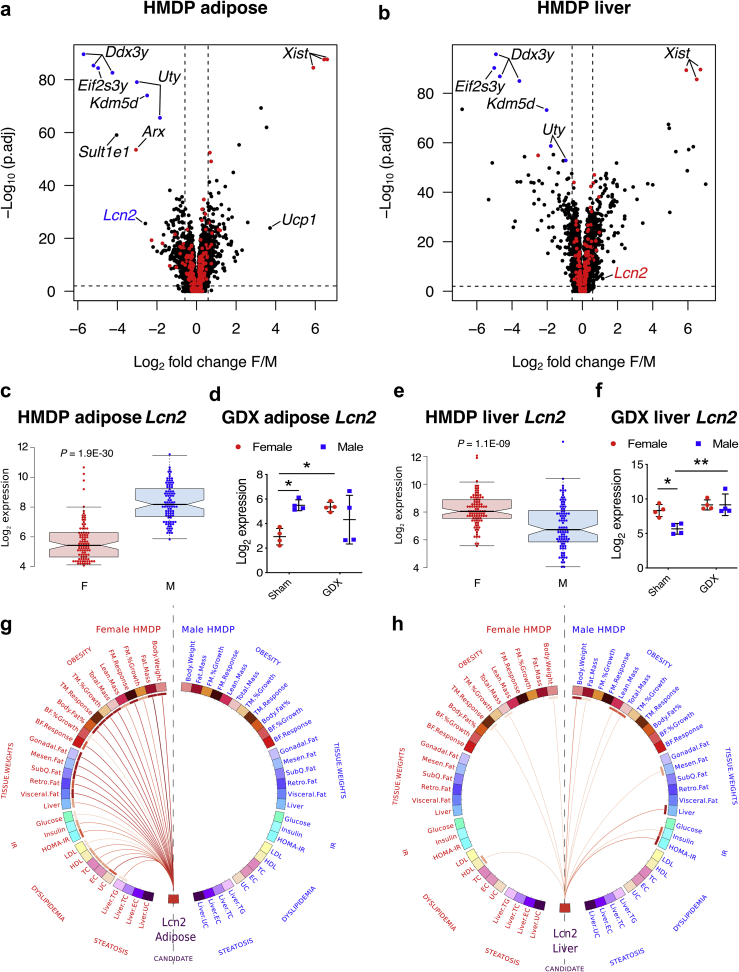
Figure 1.
Lcn2 expression in liver and adipose is highly sexually dimorphic and exhibits sex-specific associations with metabolic traits in the HMDP population, Volcano plots showing sex differences in gene expression profiles from A, adipose and B, liver tissues isolated from age- and sex-matched mouse strains (HMDP). The red and blue dots represent X and Y chromosomal genes, respectively. Average Lcn2 expression in female (red) and male (blue) C, adipose and E, liver tissues from each strain in the HF/HS fed HMDP cohort (n = 98 sex-matched strains with 2–4 mice per sex/strain). D, Adipose and F, liver Lcn2 expression in intact and gonadectomized C57BL/6J mice of both sexes (n = 4 per group). Red for females and blue for males. Circos connectogram plot showing biweight midcorrelation between G, adipose or H, liver Lcn2 expression and metabolic traits in female and male HMDP cohort (n = 102 female and 111 male strains). Each connecting line signify a significant (1% FDR cutoff) bicorrelation between Lcn2 expression and the respective trait. Red line indicates positive correlation and the color intensity signify the magnitude of correlation. Data are presented as A and B, log2 fold change (F/M); C and E, median and interquartile range; D and F, mean ± SEM and G and H, midweight bicorrelation. P values (and Q values for RNA sequencing data) were calculated by A and B, DESeq function of Bioconductor R-package for RNA sequencing; C and E, Unpaired Student's t test; D and F, 2-factor ANOVA corrected by post-hoc “Holm-Sidak's” multiple comparisons test; G and H, BicorAndPvalue function of the WGCNA R-package. *P < 0.05; **P < 0.01. See also Figures S1–S2 and Table S1.