Figure 1.
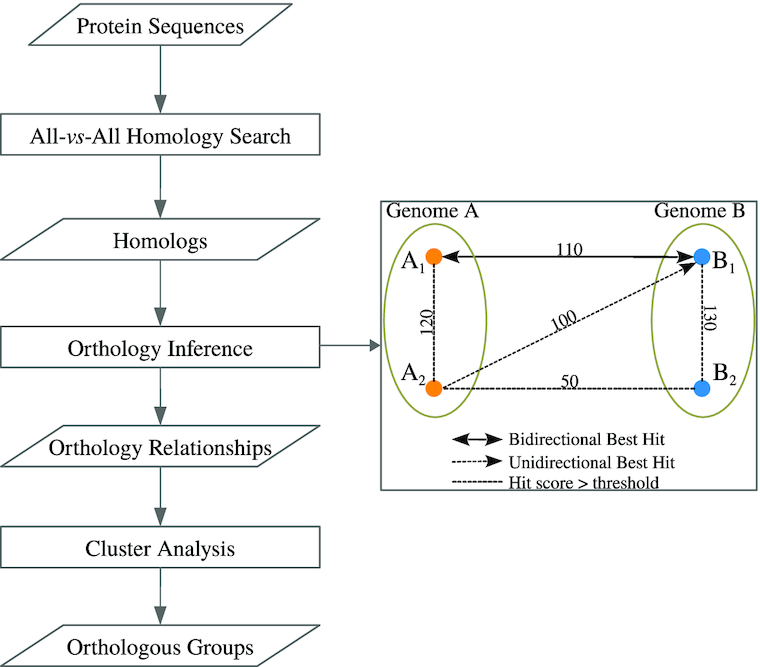
Flow chart of SwiftOrtho. SwiftOrtho is a graph-based method that consists of 3 major steps. (i) All-vs-all homology search: a seed-and-extension method is used to perform a homology search. (ii) Orthology inference: nodes are gene names; edges are similarity score of pairwise genes. 1. A1-B1 are putative orthologs identified by RBH; 2. A1-A2 and B1-B2 are putative in-paralogs because the bit scores of these pairs are greater than A1-B1; 3. A2-B1 and A2-B2 are putative co-orthologs because these pairs are not orthologs but A1-B1 are orthologs and A1-A2, B1-B2 are in-paralogs. (iii) Cluster analysis: Markov clustering or affinity propagation algorithm is used to cluster orthology relationships.
