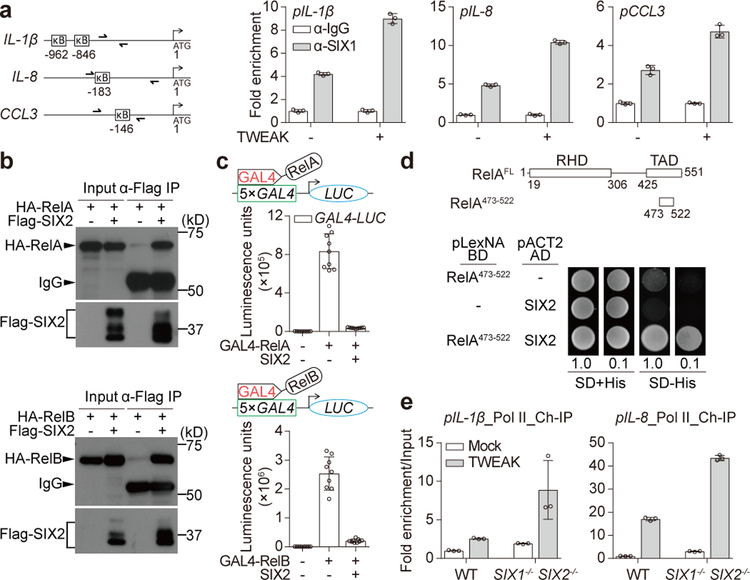
Figure 3. SIX-family transcription factors directly inhibit promoter bound NF-κB.
a, ChIP qPCR analysis of SIX1 occupancy of the indicated genes from fibroblasts treated with mock or 50 ng/ml TWEAK for 2 hours. Location of each primer set compared to the gene start sites (ATG) and κB sites are shown (diagram). IgG control samples were normalized to 1.0 and relative fold enrichment of SIX1 is shown as mean±s.d. of 3 technical replicates from one experiment. Data are representative of 3 independent experiments. b, Western blot showing input and Co-Immunoprecipitation (Co-IP) of Flag-SIX2 and association with HA-RelA (upper blot) or HA-RelB (lower blot) expressed in HEK293T cells. c, Graph showing luminescence units from 5×GAL4-Luciferase reporter gene driven by RelA (upper) and RelB (lower) fused to GAL4 DNA-binding domain. Reporter constructs were co-transfected with or without SIX2 and measured after 48 hours as indicated. Data are mean±s.d. from 9 independent experiments. d, Yeast two hybrid analysis of SIX2 binding to RelA TAD domain. Diagram shows RelA domains (RHD: Rel homology domain, TAD: Transactivation domain). Yeast transformed with the indicated plasmids were serial diluted and spotted on SD/UWL− (SD+His) or SD/WHULK− (SD-His) used to detect His-reporter gene activation by protein-protein interactions (bottom). e, The relative fold enrichment of RNA Pol II on the indicated genes in mock or 50 ng/ml TWEAK (2 hours) treatment of WT or SIX1−/− SIX2−/− fibroblasts as in a. Mock treated WT fibroblasts were normalized to 1.0 by adjusting to “input DNA” that was saved prior to immunopreciptation. Relative fold enrichment of RNA Pol II is shown as mean±s.d. of 3 technical replicates from one experiment. Data are representative of 3 independent experiments. All western blot and yeast two hybrid data are representative of 3 independent experiments. For gel source data, see Supplementary Figure 1.