Abstract
Brassica rapa includes several important leafy vegetable crops with the potential for high cadmium (Cd) accumulation, posing a risk to human health. This study aims to understand the genetic basis underlying the variation in Cd accumulation among B. rapa vegetables. Cd uptake and translocation in 64 B. rapa accessions were compared. The role of the heavy metal ATPase gene BrHMA3 in the variation of Cd accumulation was investigated. BrHMA3 encodes a tonoplast-localized Cd transporter. Five full-length and four truncated haplotypes of the BrHMA3 coding sequence were identified, explaining >80% of the variation in the Cd root to shoot translocation among the 64 accessions and in F2 progeny. Truncated BrHMA3 haplotypes had a 2.3 and 9.3 times higher shoot Cd concentration and Cd translocation ratio, respectively, than full-length haplotypes. When expressed in yeast and Arabidopsis thaliana, full-length BrHMA3 showed activity consistent with a Cd transport function, whereas truncated BrHMA3 did not. Variation in the BrHMA3 promoter sequence had little effect on Cd translocation. Variation in the BrHMA3 coding sequence is a key determinant of Cd translocation to and accumulation in the leaves of B. rapa. Strong alleles of BrHMA3 can be used to breed for B. rapa vegetables that are low in Cd in their edible portions.
Keywords: Brassica rapa, BrHMA3, cadmium, food safety, heavy metal ATPase, natural variation, translocation
Variation in the coding region of the heavy metal ATPase gene BrHMA3 controls natural variation in cadmium accumulation in widely eaten Brassica rapa vegetables.
Introduction
Cadmium (Cd) is a toxic heavy metal widely present in the environment. Cd is relatively mobile in the soil and is readily taken up by plants (Clemens et al., 2013). Contamination of agricultural soils from metal mining and smelting, industrial activities, and irrigation with waste water can further elevate Cd accumulation in food crops (Zhao et al., 2015). Moreover, widespread soil acidification due to large applications of ammonium- or urea-based nitrogen fertilizers (Guo et al., 2010) has increased Cd solubility in the soil and its uptake by crop plants (Zhao et al., 2015). Humans are exposed to Cd primarily through food consumption, with cereals and vegetables as the main dietary sources of Cd (European Food Safety Authority, 2009; Clemens et al., 2013). Depending on the dietary habits and Cd concentrations in food items, vegetables contribute 10–40% of the total dietary Cd intake (Clemens et al., 2013; He et al., 2013; Song et al., 2017; Chen et al., 2018). Due to its long biological half-life in the human body, chronic exposure to Cd can cause osteoporosis, renal dysfunction, and cancers (Jarup and Akesson, 2009; Kobayashi et al., 2009; Nordberg, 2009). In subsets of populations in China and some other Asian countries, dietary Cd intake already exceeds the tolerable limit recommended by the FAO/WHO (Meharg et al., 2013; Song et al., 2017; Wang et al., 2019). Worryingly, average dietary intake of Cd in the Chinese population has more than doubled over the last 25 years (Song et al., 2017). It is therefore imperative to reduce Cd accumulation in food crops including vegetables.
Substantial progress has been made in recent years in understanding the molecular mechanisms of Cd uptake and transport in plants, especially in Arabidopsis thaliana and rice (Clemens et al., 2013; Clemens and Ma, 2016). Cd is a hitchhiker of transporters for essential nutrients such as manganese (Mn), zinc (Zn), iron (Fe), and calcium (Ca) (Clemens, 2006). AtIRT1 (iron-regulated transporter 1), which belongs to the ZIP family (Zrt/Irt-like protein) in A. thaliana, encodes the primary transporter for Fe uptake, and also mediates Cd uptake (Connolly et al., 2002; Vert et al., 2002). The orthologues of AtIRT1 in rice, OsIRT1 and OsIRT2, have been reported to take up Cd as well as Fe (Nakanishi et al., 2006). AtNramp1 (natural resistance-associated macrophage protein 1) was reported to mediate the uptake of both Mn and Cd in A. thaliana (Cailliatte et al., 2010). In rice plants, the plasma membrane-localized OsNramp1 appears to participate in cellular Cd uptake and transport (Takahashi et al., 2011). OsNramp5 is a major influx transporter for both Mn and Cd in rice roots; knockout of this transporter leads to large decreases in the uptake of both Mn and Cd (Ishikawa et al., 2012; Ishimaru et al., 2012; Sasaki et al., 2012; Yang et al., 2014).
Plant cells have large vacuoles which play an important role in maintaining the homeostasis of nutrients and toxic elements. After Cd is taken up, a considerable portion of Cd is sequestered in the vacuoles of the root cells (Sharma et al., 2016). The P1B- ATPases as transmembrane metal transporters play an important role in metal homeostasis (Williams and Mills, 2005). The P1B-type heavy metal ATPase 3 (HMA3), a tonoplast-localized transporter, has been identified in A. thaliana and rice for transporting Cd into the vacuoles for storage (Morel et al., 2009; Ueno et al., 2010). In A. thaliana, AtHMA3 showed transport activities for Cd, Zn, Co (cobalt), and Pb (lead), and overexpression of AtHMA3 enhanced the plant tolerance to Cd (Morel et al., 2009). Based on genome-wide association studies, Chao et al. (2012) reported that natural variation in the coding sequence of AtHMA3 has a significant effect on Cd accumulation in the leaves of A. thaliana. In rice, several weak or loss-of-function alleles of OsHMA3 have been identified, which are associated with reduced Cd accumulation in the roots and increased root to shoot Cd translocation and high grain Cd concentration (Ueno et al., 2010; Miyadate et al., 2011; Yan et al., 2016; Sui et al., 2019). On the other hand, overexpression of a functional OsHMA3 gene substantially decreases the root to shoot Cd translocation in rice (Sasaki et al., 2014; Lu et al., 2019). AtHMA2 and AtHMA4 in A. thaliana and OsHMA2 in rice are involved in the loading of Cd and Zn into the xylem (Hussain et al., 2004; Verret et al., 2004; Mills et al., 2005; Wong and Cobbett, 2009; Yamaji et al., 2013). High levels of expression of AtHMA4 in the hyperaccumulator Arabidopsis halleri is an important reason for the highly efficient translocation of Cd and Zn from the roots to the shoots (Hanikenne et al., 2008). In rice, a defensin-like protein is involved in regulating the translocation of Cd to the shoots (Luo et al., 2018).
Brassica rapa includes many important vegetable crops, such as Chinese cabbage (B. rapa, ssp. pekinensis) and pak choi (B. rapa, ssp. chinensis), which are widely cultivated and consumed in Asian countries. Brassica rapa and some other vegetables in the Brassicaceae (Cruciferae) family are known to have a high ability to accumulate Cd in the edible parts (Kuboi et al., 1986), thus constituting one of the major dietary sources of Cd (Clemens et al., 2013; He et al., 2013; Chen et al., 2018). Within B. rapa, there is substantial variation in foliar Cd accumulation among different subspecies or cultivars (Liu et al., 2009, 2010; Chen et al., 2012). However, the genetic basis and the molecular mechanisms underpinning this variation are poorly understood. This knowledge is crucial for the purpose of breeding low Cd-accumulating cultivars of B. rapa, which is an important task for reducing dietary intake of Cd.
In the present study, we show that the root to shoot translocation of Cd is a critical step controlling Cd accumulation in the above-ground tissues of B. rapa, and the variation in Cd translocation and shoot Cd accumulation among 64 B. rapa accessions is largely controlled by the variation in the coding region of BrHMA3, which encodes a tonoplast-localized transporter responsible for the sequestration of Cd into the root vacuoles.
Materials and methods
Plant materials and culture conditions
Sixty-four B. rapa accessions (Supplementary Table S1 at JXB online) were obtained from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. Two F2 populations were generated from the crossing between different accessions (GJCGB×DQMY939 and GJCGB×Chiifu). After germination, seedlings were transferred to 1/10 strength Hoagland nutrient solution (pH 5.7) with aeration in a growth chamber (12 h photoperiod at 180 μM m−2 s−1 light intensity, 60% relative humidity, temperature 19–22 °C). The nutrient solution was renewed every 3 d.
Hydroponic and soil pot experiments
To investigate the variations in Cd uptake and distribution among 64 accessions of B. rapa, 18-day-old seedlings were transferred to 1/2 strength Hoagland nutrient solution containing 0.1 μM CdCl2 for 10 d, with three replicates for each accession. The solution was renewed every 3 d. After the treatment, roots were washed with 0.5 mM CaCl2 for 5 min twice. Roots and shoots were separated and rinsed with deionized water three times, before being dried at 65 °C for 3 d. Similar experiments were performed on F2 plants from the crosses of GJCGB×DQMY939 and GJCGB×Chiifu.
Nine B. rapa accessions representing nine haplotypes of BrHMA3 were selected randomly (Supplementary Table S2) and grown in an agricultural soil contaminated with a moderate level of Cd (0.95 mg Cd kg−1 soil, pH 4.7) collected from Xiangtan, Hunan province in China. One plant each of the nine accessions was grown in a plastic box containing 16 kg of soil, with four boxes representing four replicates. Plants were grown inside a growth chamber for 40 d.
Cloning of BrHMA3 and transcriptional analysis
Total RNA from the shoots and roots of B. rapa was extracted using a plant RNA extraction kit (BioTeke) and converted into cDNA using the R223-01 kit (Vazyme). The full-length coding sequence of BrHMA3 was amplified by PCR, using the primers based on the sequence of Bra024103 in the B. rapa genome database (http://brassicadb.org/brad/). The genomic fragment of BrHMA3 was cloned by PCR amplification. Bra024103 was identified as the orthologue of AtHMA3 by blasting the AtHMA3 protein sequence (Morel et al., 2009). The Bra024103 coding sequence fragments were introduced into the pEASY-Blunt vector and sequenced. The cDNA sequences and the genomic sequences were aligned by using the DNAMAN software.
BrHMA3 gene expression was determined using quantitative real-time PCR (RT-PCR) with SYBR Green reagent. The cDNA was diluted to 10 ng μl−1 and then used as the template. The actin gene (Bra005178) was used as an internal standard. The ΔCt method was used to calculate the relative expression level of BrHMA3.
Phylogenetic analysis
BrHMA3 nucleotide sequences were translated into amino acid sequences using the Primer Premier 5 software. The transmembrane domains were predicted using TOPCONS (http://topcons.net/pred/result/rst_yGKZeW/). A phylogenetic tree was constructed with the amino acid sequences of different HMAs from A. thaliana, rice, and B. rapa using the MEGA 5 software after ClustalW alignment.
Subcellular localization of BrHMA3
The full-length coding sequence of BrHMA3 (cv. Chiifu) was amplified without the stop codon. The sequence was cloned into the vector pSAT6A-eGFP-N1 (BamHI and KpnI) under the control of the Cauliflower mosaic virus 35S promoter. The 35S:BrHMA3-eGFP fragment was introduced into the expression vector pRCS2-ocs-nptII (PI-PspI). pCAMBIA-1302 vector with a 35S-eGFP (enhanced green fluorescent protein) fragment was used as a control. The constructs were transferred into Agrobacterium tumefaciens strain GV3101 and transiently expressed in tobacco (Nicotiana benthamiana) leaves (Xu et al., 2017). The protoplasts of the tobacco mesophyll cells were isolated after the removal of the cell wall by cellulase. A vector containing the AtTIP2.3 gene (At5g47450) and mCherry was co-transferred as a tonoplast-localized marker. Fluorescence of the protoplasts was observed under a confocal laser scanning microscope (LSM410; Carl Zeiss) with the excitation and emission wavelengths of 488 nm and 509 nm for GFP, and 580 nm and 610 nm for mCherry, respectively. Primers used are listed in Supplementary Table S3.
Promoter–GUS fusion construction lines
To investigate the activities of different BrHMA3 promoters, the 1100 bp promoter sequences of BrHMA3 of 10 B. rapa accessions (Supplementary Table S4) were amplified and sequenced. The promoter sequences were cloned into the PS1aG-3 vector (AscI and PacI) upstream of the GUS (β-glucuronidase) gene sequence using a ClonExpress II One Step Cloning Kit (http://www.vazyme.com/index.html). The vectors were transformed into A. thaliana (Col-0) plants using the A. tumefaciens-mediated floral dip method. To test the GUS activity, transgenic plants were immersed in GUS staining solution {50 mM phosphate-buffered saline (PBS), pH 7.0, 0.1% (w/v) Triton X-100, 10 mM Na2EDTA, 0.5 mM K3[Fe(CN)6], 0.5 mM K3[Fe(CN)6], and 1 mg ml−1 X-Gluc} at 37 °C for 6 h. The roots were photographed using a fluorescence microscope (Leica).
Yeast expression assay
Nine types of BrHMA3 coding sequences were cloned and inserted into the BamHI and KpnI sites of the pYES2 vector under the control of the GAL1 promoter. The pYES2 vector and the vector carrying the OsHMA3 gene from cv. Nipponbare were used as the negative and positive control, respectively. The constructs were transferred into yeast (Saccharomyces cerevisiae) strain INVScI. Positive clones were selected on a solid SD medium without uracil (SD-U) and cultured in a SD-U liquid medium with 2% glucose until the early log phase. Yeast cells were enriched and washed three times with sterile deionized water. The suspended cells were adjusted to an OD600 value of 1.0, and four serial dilutions were spotted on an SD-U solid medium with 2% glucose or galactose containing 0 or 7.5 μM CdCl2. The plates were cultured at 30 °C for 3 d and the growth phenotypes were observed. The growth curves of yeast cells expressing empty vector, or different BrHMA3 or OsHMA3 genes were determined in a SD-U liquid medium containing 2% galactose with or without 5 μM CdCl2. To determine Cd accumulation, yeast cells were cultured in an SD-U liquid medium containing 2% galactose with an initial OD600 of 0.2 and incubated at 30 °C for 12 h to the early log phase (OD600=0.8). CdCl2 (5 μM) was added to the medium. The cells were incubated for a further 12 h, washed with 10 mM EDTA at 4 °C three times and with deionized water three times, and then freeze-dried. The cells were digested with 5 ml of HNO3 in a microwave oven before determination of the Cd concentration.
Transgenic A. thaliana and rice expressing BrHMA3
The fragments of Types I and IX BrHMA3 coding sequences under the control of the AtHMA3 promoter were inserted into the vector pTCK303 (HindIII and SpeI). The vectors were transformed into A. thaliana (Col-0) using A. tumefaciens- (strain EHA105) mediated transformation by floral dip (Zhang et al., 2006). To generate rice expressing BrHMA3, the fragment of the Type I BrHMA3 coding sequence under the control of the maize ubiquitin promoter was inserted into the vector pTCK303 (KpnI and SpeI). The vector was transformed into rice (cv. Zhonghua 11) plants using A. tumefaciens- (strain EHA105) mediated transformation. The primers used are listed in Supplementary Table S3. The transgenic A. thaliana and rice plants were grown hydroponically and exposed to 0.5 μM CdCl2 for 3 d. Roots and shoots were harvested as described for B. rapa experiments above.
Determinations of Cd and other trace elements
Dried plant samples were ground to fine powder and digested with 5 ml of HNO3/HClO4 (85:15 v/v) in a heating block. The concentrations of Cd and other metals were determined using inductively coupled plasma mass spectrometry (ICP-MS; PerkinElmer NexION 300x).
Results
Variation in Cd accumulation among 64 Brassica rapa accessions
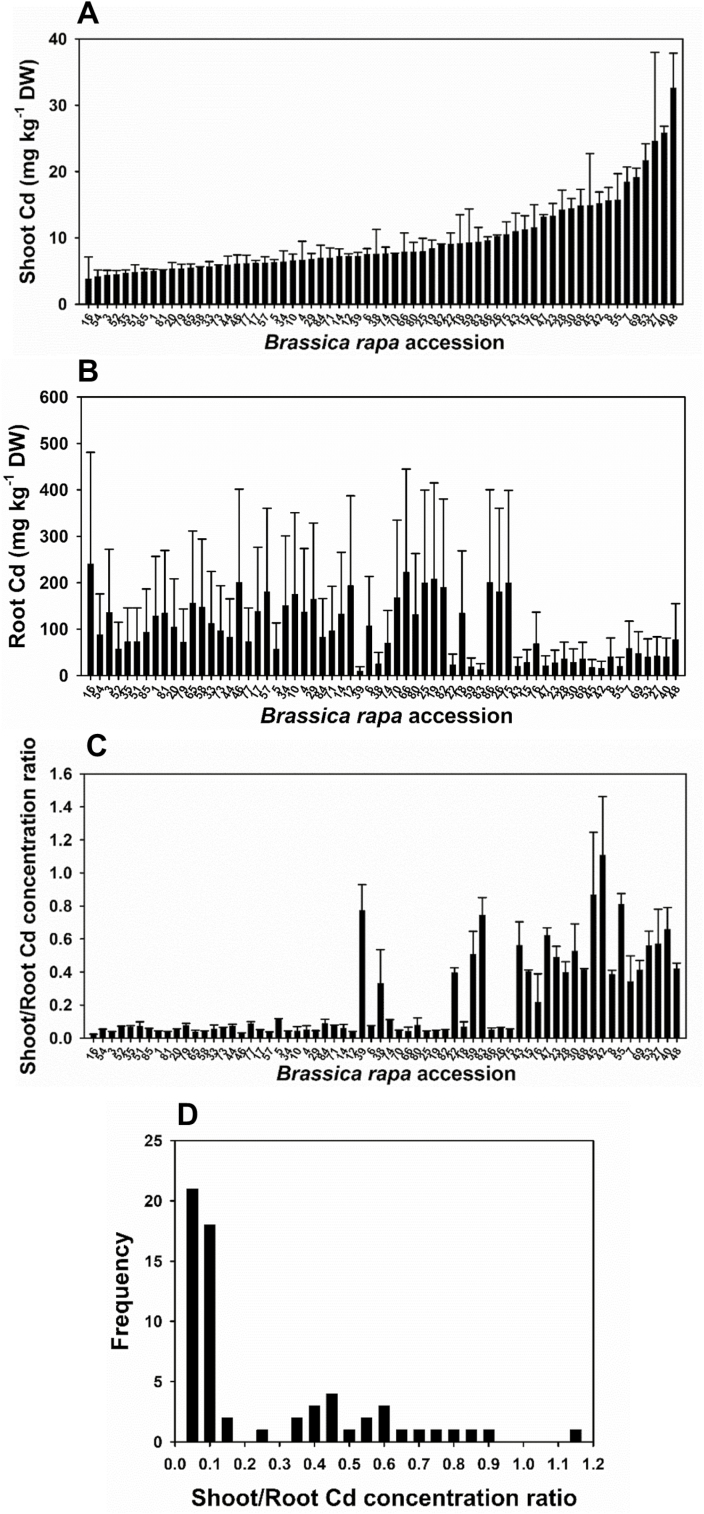
To investigate the natural variation of Cd accumulation in B. rapa, 64 accessions (Supplementary Table S1) were grown in a hydroponic culture with an environmentally relevant low concentration of Cd (0.1 μM) for 10 d. The Cd concentration in the shoots ranged from 3.8 mg kg−1 DW to 32.6 mg kg−1 DW, representing 8.5-fold variation (Fig. 1A). Root Cd concentration varied by 25-fold, from 9.6 mg kg−1 DW to 240.4 mg kg−1 DW (Fig. 1B). The shoot/root Cd concentration ratio, which is an indicator of the root to shoot translocation of Cd, varied by 47-fold (0.023–1.11) (Fig. 1C). Among the 64 accessions, this ratio appears to show a bimodal frequency distribution with a narrow group having a small ratio (<0.2) and a wide group having a large ratio (>0.2) (Fig. 1D). In general, accessions with a high Cd concentration in the shoots also have a high Cd translocation ratio.
Fig. 1.
Variation in Cd accumulation among 64 Brassica rapa accessions grown in hydroponic culture with 0.1 μM Cd. Cd concentration in the shoots (A) and roots (B), the shoot/root Cd concentration ratio (C), and frequency distribution of the Cd translocation ratio (D). Data are the means ±SD (n=3).
The concentrations of Zn, Fe, Cu, and Mn in the roots and shoots of the 64 accessions were also determined. The concentrations of Zn, Fe, Cu, and Mn in roots varied by 11.1-, 7.8-, 11.8-, and 12.3-fold, respectively (Supplementary Fig. S1), and they were not significantly correlated to the concentrations of Cd in the roots (Supplementary Fig. S2). The concentrations of Zn, Fe, Cu, and Mn in the shoots varied by 4.4-, 7.1-, 4.5-, and 2.9-fold, respectively (Supplementary Fig. S1), and there were significant correlations between the concentrations of Zn, Mn, and Cu and that of Cd in the shoots, especially between Zn and Cd (Supplementary Fig. S2).
Segregation patterns of Cd accumulation in an F2 population
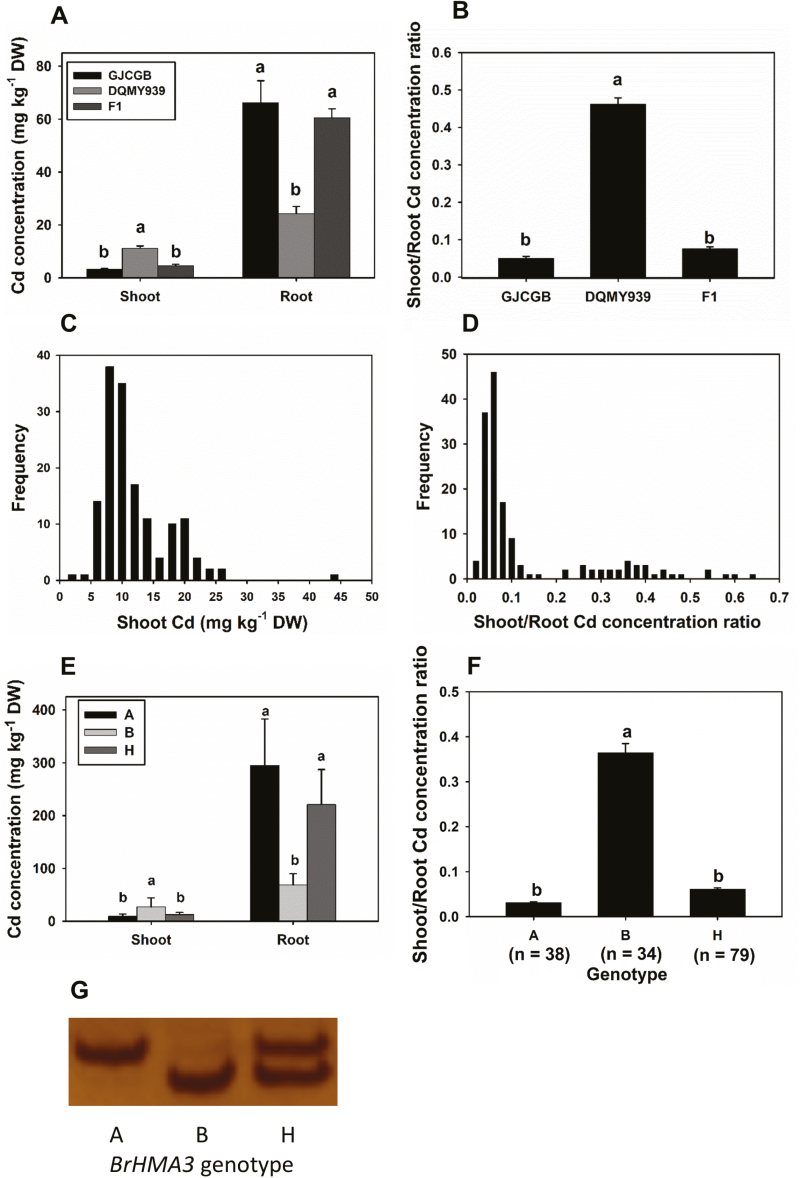
Based on the variation among the 64 B. rapa accessions, we generated an F2 segregating population from a crossing between DQMY939 (high Cd accumulation in the shoots) and GJCGB (low Cd accumulation in the shoots). When grown in hydroponic culture with 0.1 μM Cd for 10 d, GJCGB accumulated a 2.7 times higher Cd concentration in the roots but a 3.4 times lower Cd concentration in the shoots than DQMY939, with the latter having a 9.2 times higher shoot/root Cd concentration ratio than the former (Fig. 2A, B). Interestingly, the F1 plants showed a similar Cd distribution pattern to GJCGB, suggesting that the allele for high shoot Cd in DQMY939 is recessive relative to the GJCGB allele. Among the 151 F2 individuals, Cd concentrations in the shoots and roots varied by 44.8- and 11.6-fold, respectively, whilst the shoot to root Cd concentration ratio varied by 100-fold (Fig. 2C, D for shoot Cd concentration and shoot/root Cd concentration ratio, respectively). The shoot Cd concentration and, in particular, the shoot/root Cd concentration ratio showed a bimodal frequency distribution pattern (Fig. 2C, D). Using 0.2 as a separator, 118 and 33 F2 plants fell into the low and high groups of shoot/root Cd concentration ratio, respectively, which was consistent with a 3:1 segregation ratio (χ 2=0.80, df=1, P>0.05), suggesting that the Cd translocation ratio in this population is controlled by a single recessive locus.
Fig. 2.
Effects of BrHMA3 genotype on Cd accumulation in an F2 population of Brassica rapa. Cd concentrations in the roots and shoots (A) and the shoot/root Cd concentration ratio (B) of parental lines and F1 plants; frequency distributions of shoot Cd concentration (C) and the shoot/root Cd concentration ratio (D) of the F2 population; average shoot Cd concentration (E) and the shoot/root Cd concentration ratio (F) of F2 plants according to BrHMA3 genotype (G). BrHMA3 genotype: A = GJCGB, B = DQMY939, H = Heterozygous. The numbers below the genotype in (F) indicate the numbers of F2 plants in each genotype. Data are the means ±SD (n=3 in A and B, n=genotypes in E and F). Different letters above the bars represent significant differences at P<0.05 (Tukey’s test).
Isolation and polymorphism analysis of BrHMA3
It has been shown that HMA3 plays an important role in controlling Cd translocation from the roots to the shoots in A. thaliana and rice (Morel et al., 2009; Ueno et al., 2010). We therefore decided to investigate if sequence polymorphisms in BrHMA3 could explain the natural variation in Cd translocation in B. rapa. We identified the orthologue of AtHMA3 in B. rapa by Blast of the B. rapa genome (cv Chiifu) (http://brassicadb.org/brad/index.php), named BrHMA3. The gene sequence was the same as the prediction on the NCBI website with ID: XM_009139644.2. Sequence analysis showed that the ORF of BrHMA3 contains 2295 nucleotides and encodes a 764 amino acid peptide, which shows 89.15% sequence identity to AtHMA3. Phylogenetic analysis of amino acid sequences showed that BrHMA3 is closely clustered with other plant HMA3s (Supplementary Fig. S3).
The coding regions of BrHMA3 in the accessions GJCGB and DQMY939 were sequenced. BrHMA3 of GJCGB (low shoot Cd) is identical to the reference sequence in the B. rapa genome database (cv. Chiifu), whereas BrHMA3 from DQMY939 (high shoot Cd) has a premature stop codon at 1660 bp, resulting in an early translation termination at the 554th position of the protein sequence. A dCAPS (derived cleaved amplified polymorphic sequence) marker distinguishing this sequence polymorphism was developed and used for genotyping the 151 F2 individuals (Fig. 2G). The BrHMA3 genotype segregated in an ~1:2:1 ratio of BrHMA3GJCGB:heterozygotes:BrHMA3DQMY939 in the F2 population. In a hydroponic experiment with 0.1 μM Cd, the F2 plants with the BrHMA3GJCGB genotype had a mean shoot Cd concentration that was 63% lower than that in the plants with the BrHMA3DQMY939 genotype, whereas the root Cd concentration showed an opposite pattern (Fig. 2E). For the shoot/root Cd concentration ratio, the BrHMA3GJCGB genotype had a mean value of 0.031, compared with 0.36 in the plants with the BrHMA3DQMY939 genotype (Fig. 2F). For all three traits, the heterozygous plants showed a mean value that was not significantly different from that of the BrHMA3GJCGB genotype (Fig. 2E, F). Analysis using the generalized linear regression model showed that the BrHMA3 genotype explained 62% and 83% of the variation in the shoot Cd concentration and the shoot/root Cd concentration ratio, respectively, among the F2 progeny, suggesting that BrHMA3 has a major effect controlling the root to shoot translocation of Cd and Cd accumulation in the shoots of B. rapa. In contrast, the BrHMA3 genotype had no significant effect on the root and shoot Zn concentrations, or the shoot/root Zn concentration ratio (Supplementary Fig. S4). Interestingly, there were highly significant correlations between the concentrations of Zn and Cd in the shoots within each BrHMA3 genotype among the F2 population (Supplementary Fig. S4).
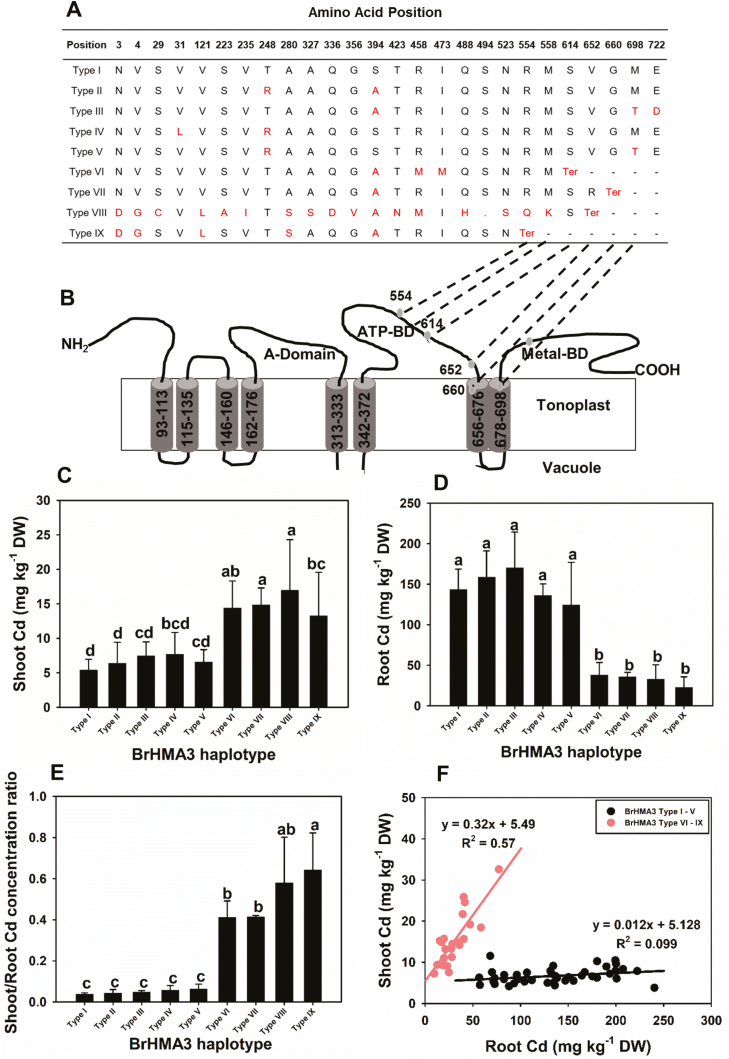
To determine the association between BrHMA3 polymorphism and Cd accumulation in the 64 B. rapa accessions, the coding regions of BrHMA3 of all accessions were cloned and sequenced. Based on the BrHMA3 protein sequences, nine haplotypes could be deduced (Fig. 3A; Supplementary Fig. S5). The frequency of each haplotype varied from 1 to 27 accessions (Supplementary Fig. S5). The haplotype of Chiifu and GJCGB is designated as Type I. Compared with Type I, Types II, IV, and V show two amino acid variations, and Type III has changes in three amino acid residues (Fig. 3A). In contrast to Types I–V, Types VI–IX all have premature terminations of protein translation at different positions, resulting in truncated proteins with 104–210 amino acid residues fewer at the C-terminus (Fig. 3A). In the truncated haplotypes, the premature terminations occur in the predicted ATP-binding domain, the seventh or the eighth transmembrane domain (Fig. 3B).
Fig. 3.
BrHMA3 protein haplotypes present in the 64 accessions of Brassica rapa and their association with Cd accumulation. Different protein haplotypes of BrHMA3 (A), predicted transmembrane domains of BrHMA3 by TOPCONS (http://topcons.cbr.su.se/) (B); mean concentrations of Cd in the shoots (C) and roots (D), and the shoot/root Cd concentration ratio (E) in 64 B. rapa accessions grouped according to BrHMA3 haplotypes; relationships between shoot and root Cd concentrations in 64 B. rapa accessions grouped according to BrHMA3 haplotypes (F). Data are the means ±SD (n=3–27). (This figure is available in colour at JXB online.)
The Cd accumulation data of the 64 accessions were then analysed according to their BrHMA3 haplotypes. The accessions possessing Types I–V had significantly lower concentrations of Cd in the shoots than those with Types VI–IX (mean difference 2.3 times), but the opposite was true for the root Cd concentrations (mean difference 4.3 times) (Fig. 3C, D). The difference in the shoot/root Cd concentration ratio was even more striking, with Types VI–IX having on average a 9.3 times higher ratio than Types I–V (Fig. 3E). Analysis based on generalized linear regression showed that BrHMA3 haplotypes explained 54% and 88% of the variation in the shoot Cd concentration and the shoot/root Cd concentration ratio, respectively. Furthermore, when the nine haplotypes were divided into two groups (group 1 for full-length Types I–V, and group 2 for truncated Types VI–IX), this simple grouping explained 52% and 87% of the variation in the shoot Cd concentration and the Cd translocation ratio, respectively. Among the 64 accessions, groups 1 and 2 contain 42 and 22 accessions, respectively (Supplementary Fig. S5). Plotting of shoot Cd concentration versus root Cd concentration reveals contrasting patterns between the accessions in these two groups. In the group 1 accessions, shoot Cd concentration hardly increased as root Cd concentration increased by >5-fold, whereas the group 2 accessions showed a linear increase in shoot Cd concentration with increasing root Cd concentration (Fig. 3F). These results indicate that, similar to the data from the F2 analysis, the polymorphism in the coding sequence of BrHMA3 is the major factor explaining the variation in the root to shoot translocation of Cd and the shoot Cd concentration among the 64 B. rapa accessions. In contrast to Cd, BrHMA3 haplotypes did not explain the variation in the concentrations of Zn, Fe, Cu, or Mn in the shoots or roots, or their shoot/root concentration ratios (Supplementary Fig. S6).
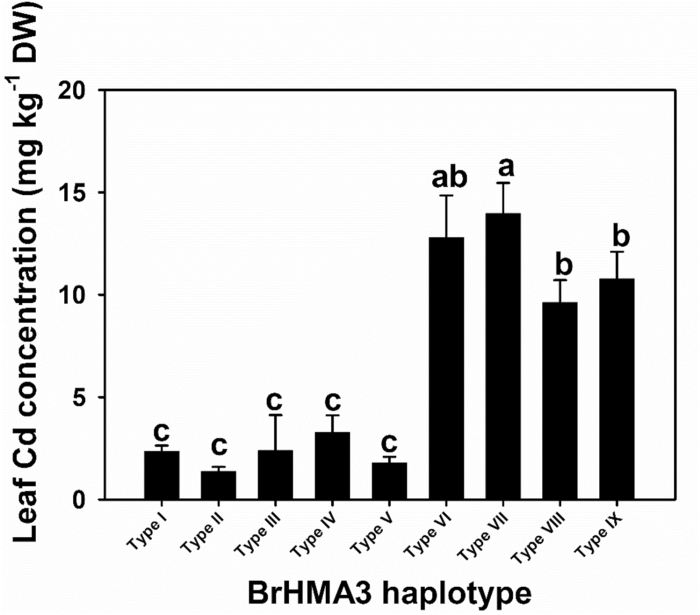
To test whether the accession difference in Cd accumulation persists in soil-grown conditions, we randomly chose one accession representing each of the nine BrHMA3 haplotypes and grew them in a Cd-contaminated soil (total Cd 0.95 mg kg−1). Consistent with the hydroponic experiment, accessions possessing Types I–V BrHMA3 had significantly lower Cd concentrations in the leaves than those of Types VI–IX, with a mean difference of 5.2-fold between the two groups (Fig. 4).
Fig. 4.
Leaf Cd concentration of Brassica rapa with different BrHMA3 haplotypes grown in a Cd-contaminated soil. Data are the means ±SD (n=3). Different letters above bars represent a significant difference at P<0.05 (Tukey’s test).
Expression pattern of BrHMA3 and subcellular localization of BrHMA3
The transcript levels of BrHMA3 in the roots and shoots of cv. Chiifu were determined using quantitative RT-PCR. BrHMA3 was mainly expressed in the roots, with very low transcript abundance in the shoots (Supplementary Fig. S7). Moreover, the expression was not induced by exposure to 5 μM Cd in hydroponic culture.
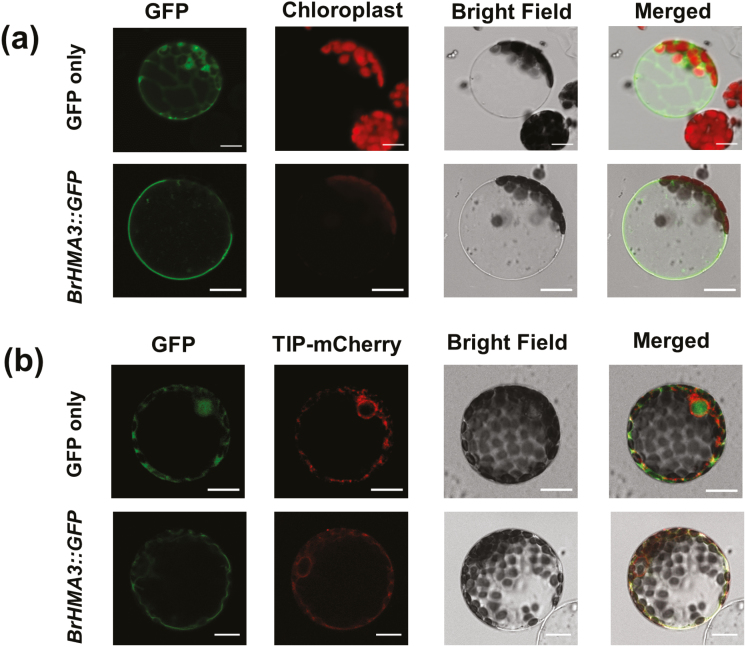
To determine the subcellular localization of BrHMA3, BrHMA3-eGFP was transiently expressed in N. benthamiana leaf mesophyll cells. The BrHMA3-fused GFP fluorescence was observed in a ring excluding the chloroplasts, suggesting a localization at the tonoplast, whereas the green fluorescence of the 35S::eGFP control could be seen throughout the cell (Fig. 5). To confirm the result, a TIP2.3-mCherry fused construct, used as a tonoplast marker, was transformed into N. benthamiana mesophyll cells together with the BrHMA3-eGFP construct (Gattolin et al., 2009). As shown in Fig. 5B, the green fluorescence of BrHMA3–eGFP coincided with the fluorescence of TIP2.3–mCherry. These results confirm that BrHMA3 is localized at the tonoplast.
Fig. 5.
Subcellular localization of BrHMA3. Protoplasts isolated from N. benthamiana leaf mesophyll cells transiently expressing BrHMA3::GFP or GFP only (A); co-transformation of TIP2.3-mCherry (a tonoplast marker) with BrHMA3::GFP or GFP (B). Scale bar=2 μm. (This figure is available in colour at JXB online.)
Cd transport activities of different BrHMA3 alleles expressed in S. cerevisiae
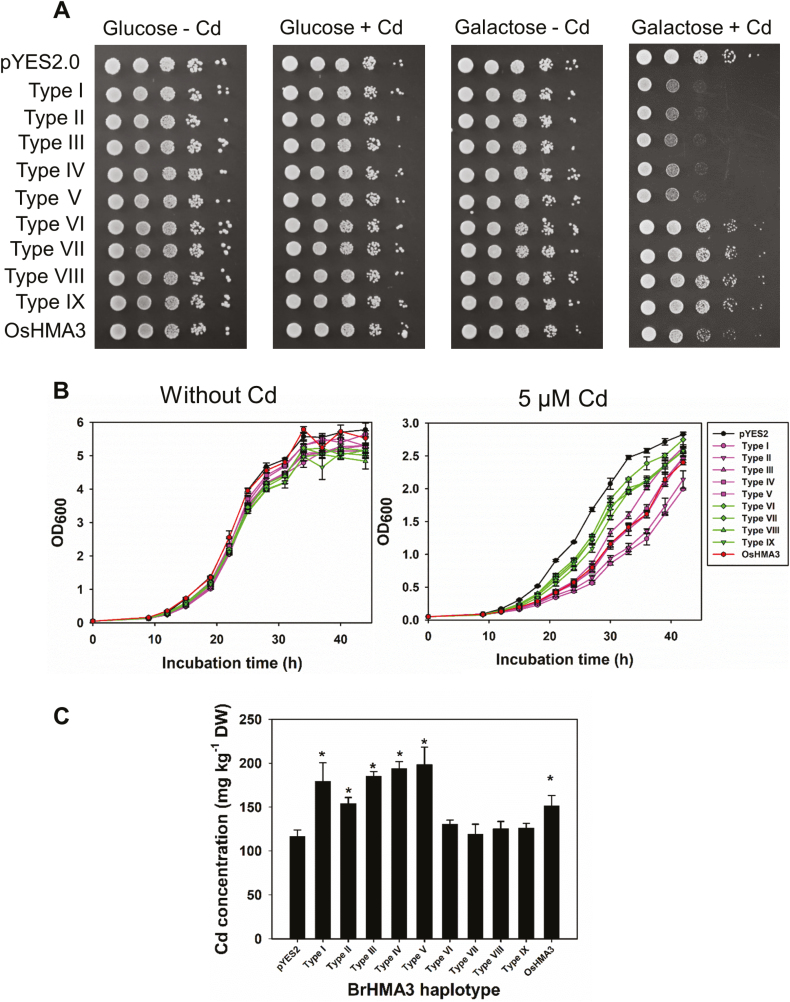
To investigate the Cd transport activity, nine different alleles of the BrHMA3 coding sequence as well as OsHMA3 from rice (cv. Nipponbare), which is known to be functional in Cd transport (Ueno et al., 2010), were fused with a GFP tag and expressed in yeast. Similar levels of GFP fluorescence intensity were observed in all the transformed yeast cells (Supplementary Fig. S8). Similar to the previous report on OsHMA3 (Ueno et al., 2010), different BrHMA3 proteins appeared to show a perinuclear localization in the yeast cells (Supplementary Fig. S8). In the presence of glucose, no difference in the Cd sensitivity was found among the yeast strains expressing either the vector control or different alleles of BrHMA3 (Fig. 6A). When galactose was supplemented as the carbon source, which induced the expression of BrHMA3, the yeast cells carrying OsHMA3 or BrHMA3 alleles coding for Types I–V showed a higher sensitivity to Cd, whereas those carrying the BrHMA3 alleles coding for Types VI–IX or empty vector showed little Cd sensitivity. Interestingly, the yeast strains expressing Types I–V BrHMA3 were even more sensitive to Cd than the yeast expressing OsHMA3 (Fig. 6A).
Fig. 6.
Functional assay of BrHMA3 through heterologous expression in yeast. Cd sensitivity tests of yeast expressing different alleles of BrHMA3, OsHMA3, or empty vector with or without Cd in the presence of glucose or galactose (A); growth curves of yeast expressing different alleles of BrHMA3, OsHMA3, or empty vector with or without Cd (B); Cd concentrations in yeast expressing different alleles of BrHMA3, OsHMA3, or empty vector after exposure to 5 μM Cd for 12 h (C). Data are the means ±SD (n=3). The asterisks above the bars in (C) indicate significant difference from the empty vector control at P<0.05 (Tukey’s test). (This figure is available in colour at JXB online.)
The growth curves of yeast cells expressing different BrHMA3 alleles were determined in the presence or absence of 5 μM Cd, with empty vector and OsHMA3 as the negative and positive control, respectively. No difference in the growth curves between the cells expressing different HMA3 genes and the vector control were observed in the absence of Cd (Fig. 6B). In the presence of 5 μM Cd, the growth of yeast cells was inhibited to different extents depending on the alleles of BrHMA3. The yeast cells expressing Types I–V BrHMA3 or OsHMA3 were inhibited more by Cd than those expressing Types VI–IX BrHMA3, the latter being slightly more inhibited than the empty vector control (Fig. 6B).
Cd accumulations in yeast cells were also determined after exposure to 5 μM Cd for 12 h. The yeast cells expressing Types I–V BrHMA3 accumulated significantly more Cd than the vector control, whereas those expressing Types VI–IX BrHMA3 accumulated similar Cd concentrations to the vector control (Fig. 6C). The results of Cd sensitivity and Cd accumulation in yeast cells suggest that Types I–V BrHMA3 have strong Cd transport activities, whereas Types VI–IX BrHMA3 had weak or no transport activities for Cd.
Expression of Type I BrHMA3 reduced Cd translocation in A. thaliana and rice
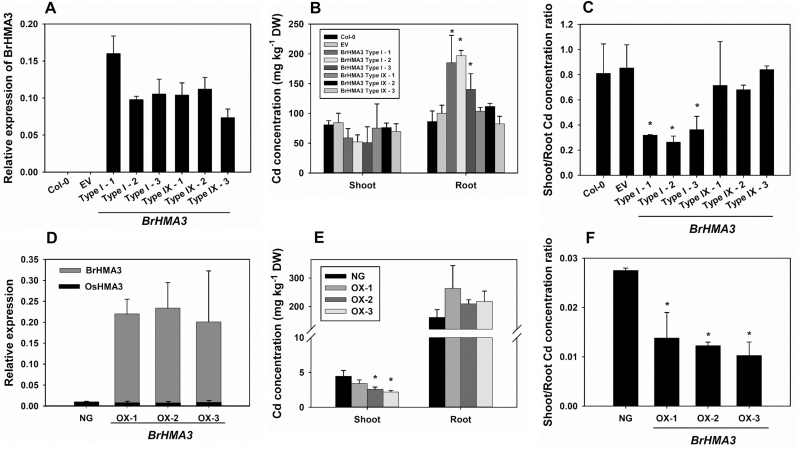
To further investigate the function of BrHAM3, we expressed Type I BrHMA3 (from cv. GJCGB) and Type IX BrHMA3 (from cv. DQMY939) in A. thaliana (Col-0) under the control of the AtHMA3 promoter from Col-0. Previous reports have shown that AtHMA3 in Col-0 is a pseudo gene (Morel et al., 2009; Chao et al., 2012). Three independent transgenic lines of each gene were selected for further characterization. In the transgenic plants, both types of BrHMA3 were mainly expressed in the roots, with similar levels of expression (Fig. 7A). In a hydroponic experiment with 0.5 μM Cd, transgenic lines of A. thaliana plants expressing Type I BrHMA3 showed a 27–85% increase in the root Cd concentration (P<0.05), but a 30–37% decrease in the shoot Cd concentration, compared with Col-0 (Fig. 7B). The shoot/root Cd concentration ratio in the Type I BrHMA3 transgenic plants was 55–61% lower than that of Col-0 (P<0.05; Fig. 7C). In contrast, transgenic plants expressing Type IX BrHMA3 showed no significant change in the root and shoot Cd concentration or the shoot/root Cd concentration ratio (Fig. 7B, C).
Fig. 7.
Functional assay of BrHMA3 through heterologous expression in Arabidopsis thaliana (A–C) and rice (D–F). Types I and IX BrHMA3 were expressed in A. thaliana (Col-0) under the control of the AtHMA3 promoter: the expression levels of BrHMA3 in the roots of transgenic plants (A); Cd concentrations in the roots and shoots (B) and the shoot/root Cd concentration ratio (C) in the wild type (Col-0), empty vector line (EV), and three independent transgenic lines. Type I BrHMA3 was expressed in rice (cv Zhonghua 11) under the control of the Ubiquitin promoter: the expression levels of BrHMA3 and OsHMA3 in the roots of a non-transgenic segregated line (NG) and three independent overexpressing lines (D); Cd concentrations in the roots and shoots (E) and the shoot/root Cd concentration ratio (F) in NG and three independent overexpressing lines. Expression of each gene was calculated as 2−ΔCT relative to AtACTIN or OsACTIN. Data are the means ±SD (n=3). Asterisks above the bars indicate significant differences from the control at P<0.05 (Tukey’s test).
We also generated transgenic rice (cv. Zhonghua11) expressing the Type I BrHMA3 gene (from cv. GJCGB) under the control of the Ubiquitin promoter. The rice cultivar used possesses a functional OsHMA3. The expression levels of BrHMA3 in the three independent transgenic lines were 22–60 times that of OsHMA3 (Fig. 7D). Compared with the segregated non-transgenic control line, the overexpressing lines showed a higher Cd concentration in the roots but lower Cd concentrations in the shoots, and a 50–63% decrease in the shoot/root Cd concentration ratio (P<0.05; Fig. 7E, F). In contrast, overexpression of BrHMA3 did not affect the Zn concentration in the shoots or the shoot to root Zn concentration ratio (Supplementary Fig. S9).
Identification of BrHMA3 promoter function
There was a substantial variation in the BrHMA3 transcript level among 10 B. rapa accessions, which differed in the root to shoot Cd translocation (Supplementary Fig. S9). To investigate whether the variation in BrHMA3 expression contributes to the variation in Cd translocation, we sequenced the promoters of BrHMA3 in the 10 accessions. The promoter sequences can be separated into two types. Type A, represented by cv. GJCGB, has a 35 bp insertion from base pair –743 to –708 and a 152 bp deletion from base pair –279 to –127 compared with the reference sequence in the cv. Chiifu (Type B) (Supplementary Fig. S10). The expression level of BrHMA3 in the roots of GJCGB was three times that of Chiifu (Supplementary Fig. S10). To investigate the activities of the two types of promoters, transgenic A. thaliana plants expressing the GUS reporter gene driven by the two types of promoters were generated. GUS staining was found in the root tips, lateral roots, and the stele region of the main roots, with the Type A promoter showing a stronger GUS activity than the Type B promoter (Supplementary Fig. S11).
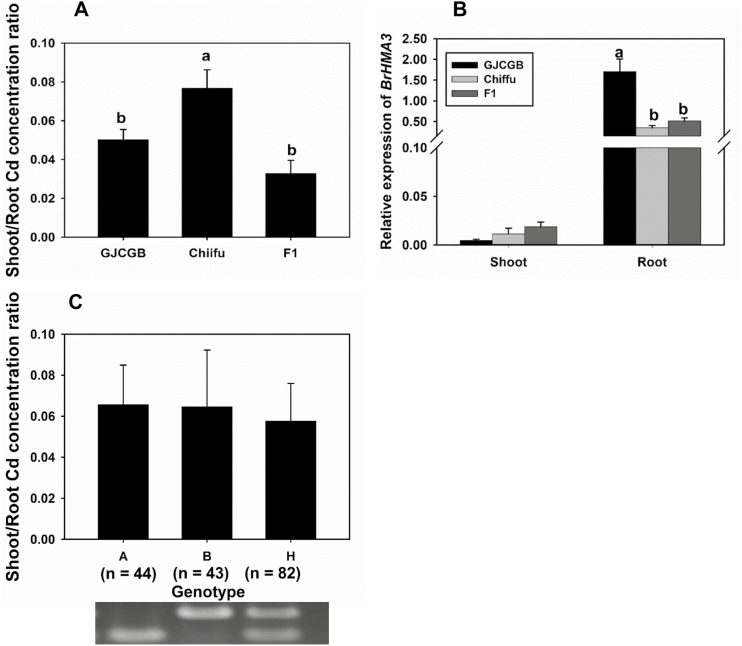
Accessions GJCGB and Chiifu have the same protein haplotype of BrHMA3 (Type I) but different promoter types. At 0.1 μM Cd, the shoot/root Cd concentration ratio in Chiifu was 1.3 times that in GJCGB (Fig. 8A). To investigate whether a difference in the promoter region of BrHMA3 could affect Cd translocation, we generated an F2 population by crossing between GJCGB and Chiifu. The F1 plants showed a similar shoot/root Cd concentration ratio to that of GJCGB (Fig. 8A), but their BrHMA3 expression level was similar to that of Chiifu (Fig. 8B). Among the 169 F2 plants, the shoot/root Cd concentration ratio varied by 8.1-fold (data not shown). The BrHMA3 genotype, determined using a marker targeting the 152 bp deletion in the promoter region, segregated approximately into a 1:2:1 ratio of PromoterGJCGB:heterozygote:PromoterChiifu among the F2 progeny (Fig. 8C). However, there was no significant difference in the shoot/root Cd concentration ratio between the three genotypes (Fig. 8C). Regression analysis using a generalized linear model showed that the genotype of the BrHMA3 promoter sequence explained only 3% of the shoot/root Cd concentration ratio among the F2 plants.
Fig. 8.
Effect of the BrHMA3 promoter on Cd accumulation in Brassica rapa. The shoot/root Cd concentration ratio in the parental lines (GJCGB and Chiifu) and F1 plants grown in hydroponic culture with 0.1 μM Cd (A); relative expression levels of BrHMA3 in roots and shoots of the parental lines (GJCGB and Chiifu) and F1 plants (B); the shoot/root Cd concentration ratio in F2 plants grouped according to BrHMA3 promoter type (C). The numbers of plants with different genotypes are indicated in (C). BrHMA3 promoter genotype: A = GJCGB, B = Chiifu, H = Heterozygous. Data are the means ±SD (n=3 in A and B). Different letters above the bars indicate significant differences at P<0.05 (Tukey’s test).
Discussion
In the present study, we show that the polymorphism of the BrHMA3 coding sequence has a pronounced effect on Cd accumulation in the edible above-ground tissues of B. rapa. This information is crucial for the purpose of breeding low Cd-accumulating cultivars of B. rapa to help reduce a significant source of dietary Cd.
BrHMA3 encodes a tonoplast-localized protein (Fig. 5). The gene is primarily expressed in the roots (Supplementary Fig. S7), a pattern that is similar to that in A. thaliana and rice. When expressed heterologously in yeast, functional alleles of BrHMA3 increased the Cd sensitivity and Cd accumulation in the yeast cells (Fig. 6), suggesting that BrHMA3 is able to transport Cd. The increased Cd sensitivity and accumulation were probably caused by the mislocalization of BrHMA3 to the organelles such as the endoplasmic reticulum in the yeast cells, in addition to the vacuoles (Supplementary Fig. S8). Similar observations were found when OsHMA3 and SpHMA3 were expressed in yeast (Ueno et al., 2010; Liu et al., 2017). When a functional allele of BrHMA3 was expressed in A. thaliana and rice, it had the effect of decreasing Cd translocation from the roots to the shoots (Fig. 7). These results indicate that BrHMA3, like its homologues AtHMA3 and OsHMA3 (Morel et al., 2009; Ueno et al., 2010; Chao et al., 2012), functions as a tonoplast-localized heavy metal ATPase capable of transporting Cd into the vacuoles in the root cells and hence limiting Cd translocation to the shoot tissues.
We found a large variation in Cd accumulation in the shoots of 64 accessions of B. rapa (Fig. 1). This variation is primarily related to the variation in the root to shoot translocation of Cd. Similarly, the root to shoot translocation of Cd is a key determinant of Cd accumulation in rice cultivars (Uraguchi et al., 2009). Because variations in AtHMA3 and OsHMA3 coding regions are known to affect Cd translocation in A. thaliana and rice, respectively (Ueno et al., 2010; Chao et al., 2012; Yan et al., 2016; Sui et al., 2019), we analysed the coding sequences of BrHMA3 in the 64 accessions of B. rapa. Nine protein haplotypes of BrHMA3 were found, including five full-length and four truncated sequences. The results from heterologous expression in yeast and A. thaliana support the notion that the full-length haplotypes of BrHMA3 have Cd transport activity, whereas the truncated BrHMA3 are null alleles with regard to Cd transport (Figs 6, 7). There was a strong association between the BrHMA3 haplotypes and the phenotypes of Cd translocation from the roots to the shoots and shoot Cd concentration among the 64 accessions (Fig. 3). Strikingly, accessions with truncated BrHMA3 haplotypes had ~10 times higher shoot/root Cd concentration ratios than those with the full-length haplotypes. Furthermore, in an F2 population from a crossing between two accessions possessing Types I and IX BrHMA3, the BrHMA3 genotype explained a large proportion of the variation in the Cd translocation and accumulation in the shoots (Fig. 2). These results provide genetic evidence that BrHMA3 is a key determinant of Cd translocation to and accumulation in the shoots of B. rapa through its effect on Cd sequestration in the vacuoles of root cells.
Whilst the variation in the coding region of BrHMA3 affects the transport activity for Cd, variation in the expression level may also play a role, as has been shown for natural variation in Na and Mo accumulation being driven by expression level polymorphisms in HKT1, AtMOT1, and OsMOT1;1, respectively (Tomatsu et al., 2007; Baxter et al., 2008, 2010; Huang et al., 2019). To test this hypothesis, we generated an F2 population from a crossing between two accessions possessing the same full-length coding sequence of BrHMA3 (haplotype Type I) but having different promoter sequences with different promoter activities (Fig. 8; Supplementary Fig. S10). However, variation in the BrHMA3 promoter among the F2 individuals had no significant effect on Cd translocation (Fig. 8), suggesting that, in contrast to the coding region variation, the variation in the BrHMA3 promoter is not a key determinant of Cd translocation. It is possible that the variation in the expression level of BrHMA3 is not large enough to cause a significant effect on Cd sequestration and translocation. Alternatively, it is also possible that other genes contribute to the relatively small variation in Cd translocation between the parental lines, thus masking the effect of promoter variation.
AtHMA3 and OsHMA3 can transport Zn in addition to Cd (Morel et al., 2009; Sasaki et al., 2014). Whether BrHMA3 can transport Zn was not tested directly in our study. However, the coding sequence polymorphism in BrHMA3 did not affect Zn translocation to and accumulation in the shoots (Supplementary Figs S4, S6), and overexpression of BrHMA3 in rice did not affect Zn translocation or accumulation (Supplementary Fig. S9). One possibility is that Zn homeostasis is tightly regulated, and the effect of BrHMA3, if any, may be masked by other genes. We did observe strong correlations between the concentrations of Zn and Cd in the shoots among the 64 B. rapa accessions (Supplementary Fig. S2) and, in particular, in a segregating F2 population, which was independent of the BrHMA3 genotype (Supplementary Fig. S4). These results suggest that there probably exist other genes that control the uptake and/or the translocation of Zn and Cd simultaneously in B. rapa.
It is interesting that natural variation in the HMA3 coding sequences controlling Cd translocation and accumulation in the shoots is a conserved mechanism across plant species, including A. thaliana (Chao et al., 2012), rice (Ueno et al., 2010; Yan et al., 2016; Sui et al., 2019), and B. rapa (the present study). However, it is unclear how different alleles of HMA3 are selected under the natural environment. One possibility is that strong alleles of HMA3 may have an adaptive advantage in high Cd soils because vacuolar sequestration is an important mechanism of Cd detoxification. This possibility may be discounted because soil contamination with Cd, although an important issue for food safety, is usually not high enough to cause toxicity to plants, which is contrary to the strong BrHMA3 alleles being more frequent than the weak alleles among the B. rapa accessions (Supplementary Fig. S5). Alternatively, selection on HMA3 alleles may act through their effect on the sequestration of essential elements, such as Zn, in that weak alleles could be beneficial in allowing more Zn to be accumulated in the shoots (Chao et al., 2012; Sasaki et al., 2014). When a strong AtHMA3 allele was expressed with its native promoter in A. thaliana Col-0, which possesses a null allele of AtHMA3, a small but significant decrease in leaf Zn concentration was observed (Chao et al., 2012), supporting the notion that selection on AtHMA3 is manifested through its role in Zn homeostasis. Whether BrHMA3 alleles are selected through the Zn homeostasis effect remains unknown.
In conclusion, we show that BrHMA3 is a key gene controlling Cd translocation to and accumulation in the edible parts of B. rapa vegetables. Allelic variation in the coding region of BrHMA3 largely explains the variation in Cd translocation to and accumulation in the shoots among different B. rapa accessions. Haplotypes of BrHMA3 with strong Cd transport activities identified in the present study will be very useful for breeding low Cd-accumulating accessions of B. rapa vegetables through molecular marker-assisted breeding programmes. Null alleles of BrHMA3, probably associated with high Cd accumulation in the shoots, can also be screened in existing accessions using appropriate molecular markers. Given the 5-fold difference in the leaf Cd concentration between the functional and null alleles when different accessions were grown in soil (Fig. 4) and the high consumption of B. rapa vegetables, a simple screening based on the BrHMA3 genotype could make a significant contribution to reducing the dietary intake of Cd.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Concentrations of Zn, Fe, Mn, and Cu in 64 Brassica rapa accessions.
Fig. S2. Correlations between Cd and other trace metals.
Fig. S3. Phylogenetic analysis of HMA3 genes.
Fig. S4. Zinc accumulation in GJCGB×DQMY939 F2 plants.
Fig S5. Amino acid sequence alignment of nine BrHMA3 protein haplotypes.
Fig. S6. Effects of BrHMA3 haplotypes on Zn, Mn, Fe, Cu, and Cd accumulation.
Fig. S7. BrHMA3 expression levels in Brassica rapa roots and shoots.
Fig. S8. Intracellular localization of GFP–BrHMA3 expressed in yeast.
Fig. S9. Zn concentration in BrHMA3 transgenic rice plants.
Fig. S10. Cd accumulation and BrHMA3 expression levels in 10 Brassica rapa accessions.
Fig. S11. Analysis of the BrHMA3 promoter activity.
Table S1. List of 64 Brassica rapa accessions used in the present study.
Table S2. Nine Brassica rapa accessions used in the soil pot experiment.
Table S3. Primers used in the present study.
Table S4. Ten Brassica rapa accessions selected for promoter analysis.
Acknowledgements
This work was supported by the Natural Science Foundation of China (31520103914), the Innovative Research Team Development Plan of the Ministry of Education of China (grant no. IRT_17R56), and the Fundamental Research Funds for the Central Universities (grant no. KYT201802).
References
- Baxter I, Brazelton JN, Yu D, et al. . 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genetics 6, e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Muthukumar B, Park HC, et al. . 2008. Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genetics 4, e1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. 2010. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. The Plant Cell 22, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE. 2012. Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genetics 8, e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yang X, Wang P, Wang Z, Li M, Zhao FJ. 2018. Dietary cadmium intake from rice and vegetables and potential health risk: a case study in Xiangtan, southern China. The Science of the Total Environment 639, 271–277. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li TQ, Han X, Ding ZL, Yang XE, Jin YF. 2012. Cadmium accumulation in different pakchoi cultivars and screening for pollution-safe cultivars. Journal of Zhejiang University. Science. B 13, 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. 2006. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719. [DOI] [PubMed] [Google Scholar]
- Clemens S, Aarts MG, Thomine S, Verbruggen N. 2013. Plant science: the key to preventing slow cadmium poisoning. Trends in Plant Science 18, 92–99. [DOI] [PubMed] [Google Scholar]
- Clemens S, Ma JF. 2016. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annual Review of Plant Biology 67, 489–512. [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. 2002. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell 14, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority 2009. Scientific opinion of the panel on contaminants in the food chain on a request from the European Commission on cadmium in food. EFSA Journal 980, 1–139. [Google Scholar]
- Gattolin S, Sorieul M, Hunter PR, Khonsari RH, Frigerio L. 2009. In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biology 9, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KW, Vitousek PM, Zhang FS. 2010. Significant acidification in major Chinese croplands. Science 327, 1008–1010. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. 2008. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453, 391–395. [DOI] [PubMed] [Google Scholar]
- He P, Lu Y, Liang Y, Chen B, Wu M, Li S, He G, Jin T. 2013. Exposure assessment of dietary cadmium: findings from Shanghainese over 40 years, China. BMC Public Health 13, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Liu H, Zhu YF, Pinson SRM, Lin HX, Guerinot ML, Zhao FJ, Salt DE. 2019. Natural variation in a molybdate transporter controls grain molybdenum concentration in rice. New Phytologist 221, 1983–1997. [DOI] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. 2004. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. The Plant Cell 16, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa NK, Nakanishi H. 2012. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proceedings of the National Academy of Sciences, USA 109, 19166–19171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Takahashi R, Bashir K, et al. . 2012. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Scientific Reports 2, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järup L, Akesson A. 2009. Current status of cadmium as an environmental health problem. Toxicology and Applied Pharmacology 238, 201–208. [DOI] [PubMed] [Google Scholar]
- Kobayashi E, Suwazono Y, Dochi M, Honda R, Kido T. 2009. Influence of consumption of cadmium-polluted rice or Jinzu River water on occurrence of renal tubular dysfunction and/or Itai-itai disease. Biological Trace Element Research 127, 257–268. [DOI] [PubMed] [Google Scholar]
- Kuboi T, Noguchi A, Yazaki J. 1986. Family dependent cadmium accumulation characteristics in higher plants. Plant and Soil 92, 405–415. [Google Scholar]
- Liu H, Zhao H, Wu L, Liu A, Zhao FJ, Xu W. 2017. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytologist 215, 687–698. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou Q, An J, Sun Y, Liu R. 2010. Variations in cadmium accumulation among Chinese cabbage cultivars and screening for Cd-safe cultivars. Journal of Hazardous Materials 173, 737–743. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou Q, Sun Y, Liu R. 2009. Identification of Chinese cabbage genotypes with low cadmium accumulation for food safety. Environmental Pollution 157, 1961–1967. [DOI] [PubMed] [Google Scholar]
- Lu C, Zhang L, Tang Z, Huang XY, Ma JF, Zhao FJ. 2019. Producing cadmium-free Indica rice by overexpressing OsHMA3. Environment International 126, 619–626. [DOI] [PubMed] [Google Scholar]
- Luo JS, Huang J, Zeng DL, et al. . 2018. A defensin-like protein drives cadmium efflux and allocation in rice. Nature Communications 9, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg AA, Norton G, Deacon C, et al. . 2013. Variation in rice cadmium related to human exposure. Environmental Science & Technology 47, 5613–5618. [DOI] [PubMed] [Google Scholar]
- Mills RF, Francini A, Ferreira da Rocha PS, Baccarini PJ, Aylett M, Krijger GC, Williams LE. 2005. The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Letters 579, 783–791. [DOI] [PubMed] [Google Scholar]
- Miyadate H, Adachi S, Hiraizumi A, et al. . 2011. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytologist 189, 190–199. [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. 2009. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiology 149, 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK. 2006. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Science and Plant Nutrition 52, 464–469. [Google Scholar]
- Nordberg GF. 2009. Historical perspectives on cadmium toxicology. Toxicology and Applied Pharmacology 238, 192–200. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF. 2014. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. Journal of Experimental Botany 65, 6013–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF. 2012. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. The Plant Cell 24, 2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SS, Dietz KJ, Mimura T. 2016. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant, Cell & Environment 39, 1112–1126. [DOI] [PubMed] [Google Scholar]
- Song Y, Wang Y, Mao W, Sui H, Yong L, Yang D, Jiang D, Zhang L, Gong Y. 2017. Dietary cadmium exposure assessment among the Chinese population. PLoS One 12, e0177978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui F, Zhao D, Zhu H, Gong Y, Tang Z, Huang XY, Zhang G, Zhao FJ. 2019. Map-based cloning of a new total loss-of-function allele of OsHMA3 causes high cadmium accumulation in rice grain. Journal of Experimental Botany 70, 2857–2871. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK. 2011. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. Journal of Experimental Botany 62, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatsu H, Takano J, Takahashi H, Watanabe-Takahashi A, Shibagaki N, Fujiwara T. 2007. An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proceedings of the National Academy of Sciences, USA 104, 18807–18812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF. 2010. Gene limiting cadmium accumulation in rice. Proceedings of the National Academy of Sciences, USA 107, 16500–16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S. 2009. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. Journal of Experimental Botany 60, 2677–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. 2004. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Letters 576, 306–312. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Chen H, Kopittke PM, Zhao FJ. 2019. Cadmium contamination in agricultural soils of China and the impact on food safety. Environmental Pollution 249, 1038–1048. [DOI] [PubMed] [Google Scholar]
- Williams LE, Mills RF. 2005. P(1B)-ATPases—an ancient family of transition metal pumps with diverse functions in plants. Trends in Plant Science 10, 491–502. [DOI] [PubMed] [Google Scholar]
- Wong CKE, Cobbett CS. 2009. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytologist 181, 71–78. [DOI] [PubMed] [Google Scholar]
- Xu J, Shi S, Wang L, Tang Z, Lv T, Zhu X, Ding X, Wang Y, Zhao FJ, Wu Z. 2017. OsHAC4 is critical for arsenate tolerance and regulates arsenic accumulation in rice. New Phytologist 215, 1090–1101. [DOI] [PubMed] [Google Scholar]
- Yamaji N, Xia J, Mitani-Ueno N, Yokosho K, Feng Ma J. 2013. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiology 162, 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang P, Wang P, Yang M, Lian X, Tang Z, Huang CF, Salt DE, Zhao FJ. 2016. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant, Cell & Environment 39, 1941–1954. [DOI] [PubMed] [Google Scholar]
- Yang M, Zhang Y, Zhang L, et al. . 2014. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. Journal of Experimental Botany 65, 4849–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Ma Y, Zhu YG, Tang Z, McGrath SP. 2015. Soil contamination in China: current status and mitigation strategies. Environmental Science & Technology 49, 750–759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.