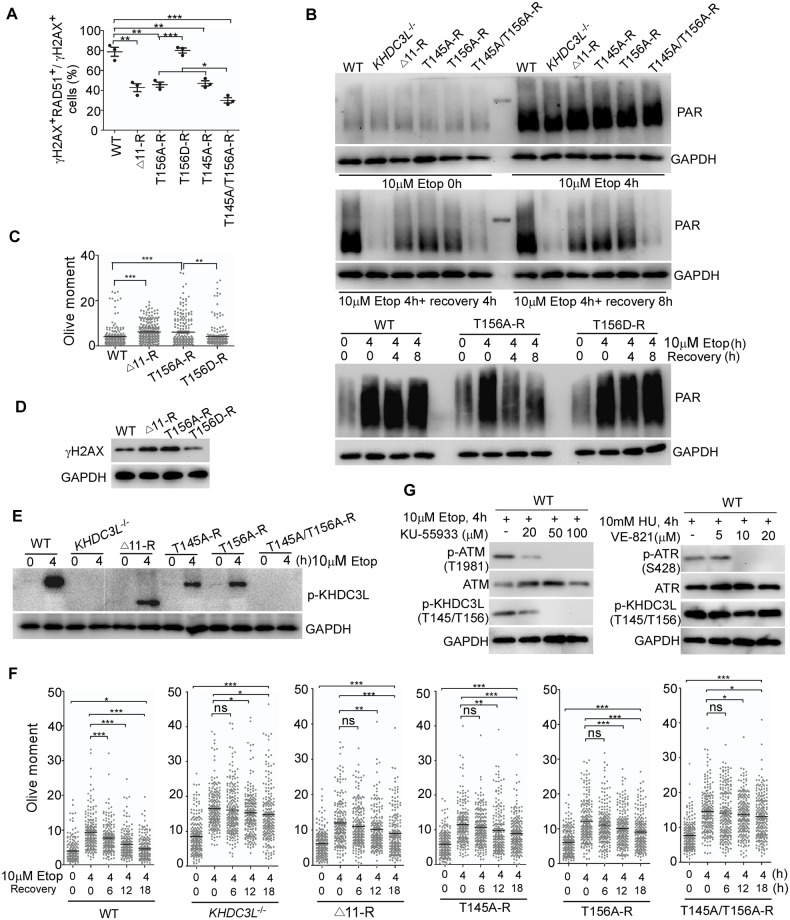
Fig 7. Phosphorylation of T156 and T145 sites in KHDC3L by ATM is required for HR repair and PARP1 activation.
(A) T156A, T145A, and Δ11 mutations caused similar level of defect in recruiting RAD51 to DNA DSB sites, whereas T156D had no effects on RAD51 recruitment. Double mutations of T156A/T145A displayed a more severe phenotype than those of individual mutation (n = 50 in one replicate, total three independent experiments). (B) PARP1 activity in hESCs with WT or mutant KHDC3L proteins. Neutral comet assay (C) (n = 200 from two independent experiments) and immunoblotting (D) confirmed that hESCs expressing Δ11 or T156A mutant KHDC3L accumulated a higher level of DNA DSBs than did WT hESCs or ESCs expressing T156D. (E) Phosphorylation of T145/T156 sites at different hESC lines. (F) T145A, T156A, and Δ11 mutations displayed similar level of defects in DSB repair (n = 200 from two independent experiments). Similar to KHDC3L−/−, T156A/T145A double mutations caused more-severe defects. (G) Complete repression of ATM activation by KU-55933 eliminated the phosphorylation of T145/T156 in KHDC3L, whereas inhibition of ATR activity by VE-821 had no effects. Student two-tailed t test was performed for statistical analysis. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Underlying numerical values in (A), (C), and (F) can be found in S1 Data. Δ11, p.E150_V160del; ATM, Ataxia-telangiectasia mutated; ATR, Ataxia-telangiectasia and Rad3-related protein; DSB, double-strand break; ESC, embryonic stem cell; Etop, etoposide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; hESC, human ESC; HR, homologous recombination; KHDC3L, KH domain containing 3 like; ns, not significant; PAR, poly(ADP-ribose); PARP, PAR polymerase; RAD51, RAS associated with diabetes protein 51; WT, wild type.