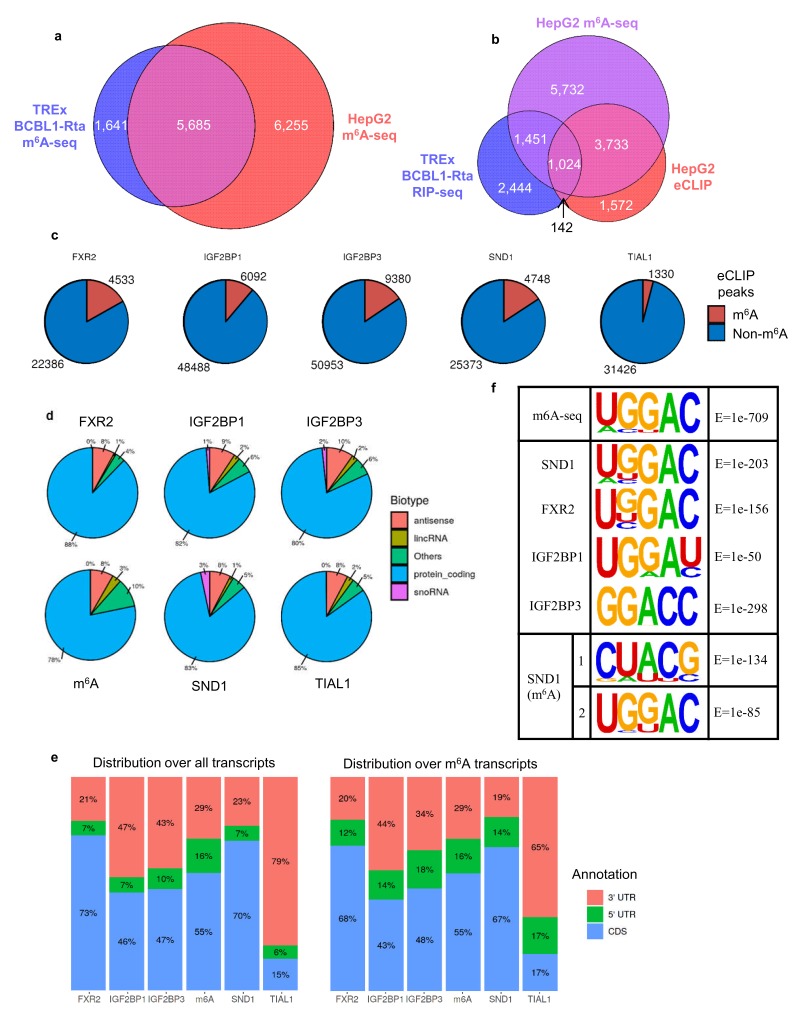
Figure 6. SND1 binds the consensus m6A motif.
(a) Overlap of m6A-modified transcripts in the indicated cell lines. (b) Overlap between the indicated eCLIP, RIP-seq and m6A-seq enriched transcripts. (c) Direct overlap of m6A-seq peaks and eCLIP binding sites for m6A reader proteins (FXR2, IGF2BP1, IGF2BP3), SND1 and the control, TIAL1. (d) Distribution of binding sites for the indicated proteins across RNA biotypes. (e) Distribution of binding sites across different transcript regions for the indicated proteins. (f) Motif analysis using HOMER and HepG2 eCLIP datasets as indicated. Top panel m6A-seq derived motif in HepG2 cells (note that this is the highest scoring motif derived), central panels are consensus motifs found using all target transcripts, bottom panel is the two most highly enriched SND1 motifs in the set of m6A-modified exons bound by SND1.