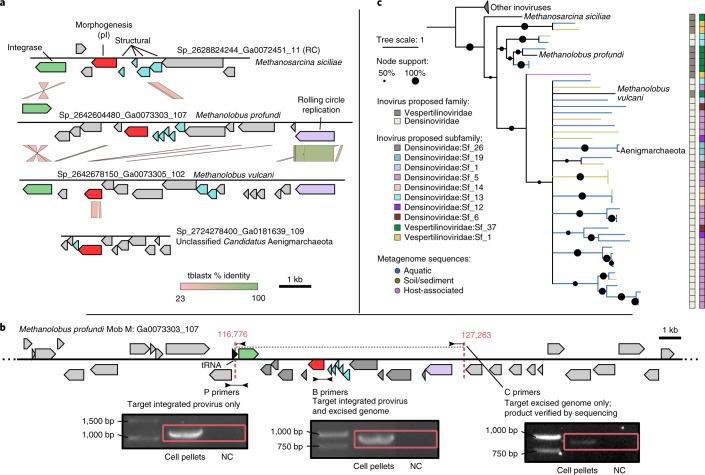
Fig. 4. Characterization of archaea-associated inoviruses.
a, Genome comparison of the four inovirus sequences detected in members of the Methanosarcinaceae family or Aenigmarchaeota candidate phylum. Genes are coloured according to their functional affiliation (light grey indicates ORFan). RC, sequence is reverse complemented. b, PCR validation of the predicted inovirus from the archaea host M. profundi MobM. Three primer pairs were designed and used to amplify across the predicted 5′ insertion site (P primers), within the predicted provirus (B primers) or across the junction of the predicted excised circular genome (C primers). The predicted provirus attachment site is indicated by dotted red lines along with corresponding genome coordinates. Products from C primers were sequenced and aligned to the M. profundi MobM genome to confirm that they spanned both ends of the provirus in the expected orientation and at the predicted coordinates (see Supplementary Notes and Supplementary Fig. 6). Red boxes indicate the expected product lengths. P and B primer amplifications were repeated twice, and the C primer amplifications were repeated three times, with an identical result obtained for each replicate (Supplementary Fig. 11). NC, no template control. c, Phylogenetic tree of archaea-associated inoviruses and related sequences. The tree was built from pI protein multiple alignment with IQ-TREE. Nodes with support of <50% were collapsed. Branches leading to inovirus species associated to a host are coloured in black, and the corresponding host is indicated on the tree. Branches leading to inovirus species assembled from metagenomes are coloured by type of environment. Classification of each inovirus species in proposed families and subfamilies is indicated next to the tree (see Fig. 5).