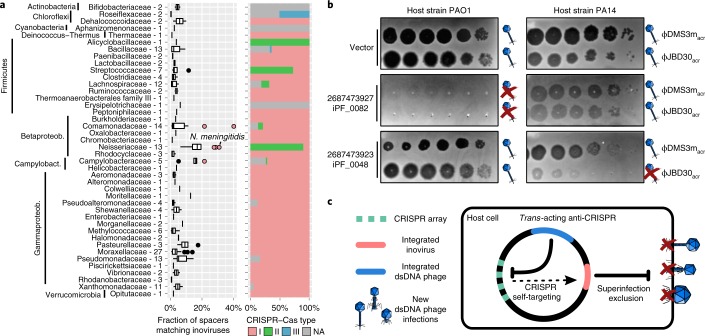
Fig. 6. Interaction of inoviruses with CRISPR–Cas systems and co-infecting viruses.
a, Proportion of the spacers matching an inovirus genome and the corresponding distribution of CRISPR–Cas systems. The proportions are calculated only on hosts with at least one spacer matching an inovirus sequence, with hosts grouped at the family rank (hosts unclassified at this rank were not included). In the boxplot, the lower and upper hinges correspond to the first and third quartiles, respectively, and the whiskers extend no further than ±1.5 times the interquartile range. Outliers identified as values larger than the third quartile plus three times the interquartile range from the complete distribution are highlighted in red. The number of observations is indicated next to each family. b, Instances of superinfection exclusion observed when expressing individual inovirus genes in two P. aeruginosa strains: PAO1 and PA14. From top to bottom: cells were transformed with an empty vector, one expressing gene 2687473927 or one expressing gene 2687473923. For each construct, host cells were challenged with serial dilutions (from left to right) of phages: ϕJBD30 and ϕDMS3m. The formation of plaques (dark circles) indicates successful infection, whereas the absence of plaques indicates superinfection exclusion. Interpretation of infection outcome is indicated to the right of each lane, with successful infection represented by a phage symbol and superinfection exclusion represented by a phage symbol barred by a red cross. Results from additional superinfection exclusion experiments are presented in Supplementary Figs. 10 and 12. All superinfection experiments were conducted twice and produced similar results. c, Schematic representation of the possible mutualistic or antagonistic interactions between inovirus prophages (red) and co-infecting Caudovirales (blue). Mutualistic interactions include suppression of the CRISPR–Cas immunity, especially for integrated inoviruses targeted by the host cell CRISPR–Cas system (‘self-targeting’). Antagonistic interactions primarily involve superinfection exclusion, in which a chronic inovirus infection prevents a secondary infection by an unrelated virus.