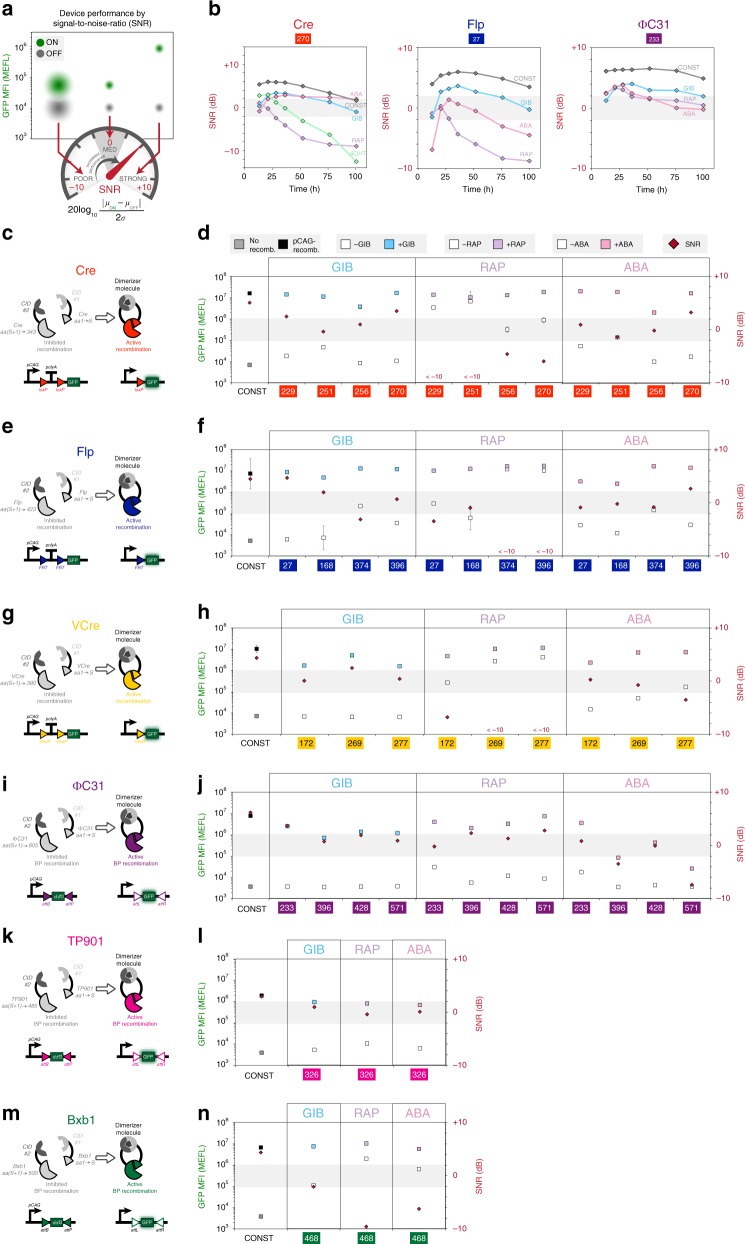
Fig. 2.
Swapping chemical-inducible dimerization domains yields a large repertoire of small-molecule inducible recombinases. a A signal-to-noise (SNR) metric can be used to capture distinguishability of on and off states, accounting for both the absolute difference in mean signal expression and spread (noise) of the distributions. SNR dynamics in units of decibels (dB) were captured over 100 h (b) post transfection for particular amino acid splits of inducible Cre, Flp, and ɸC31 systems incorporating the gibberellin (GIB), rapalog (RAP), and abscisic acid (ABA) dimerization domains GAI/GID1, FKBP/FRB, and PYL/ABI, respectively. A 4-hydroxytamoxifen (4OHT)-inducible pCAG-ERT2-Cre-ERT2 construct is also included in b. Various splits of Cre (c, d), Flp (e, f), VCre (g, h), ɸC31 (i, j), TP901 (k, l), and Bxb1 (m, n) were incorporated with the GIB-, RAP-, and ABA-associated dimerization domains. Measurements of GFP mean fluorescence intensity (MFI) in units of molecules of equivalent fluorescein (MEFL) and SNR were made for plus drug (colored blue, purple, and pink squares corresponding to gibberellin, rapalog, and abscisic acid added 2 h post transfection) and minus drug (white squares) conditions. Constitutive recombinases (black squares) and blank vectors (gray squares) were also transfected as a comparison. Red diamonds indicate SNR score. Error bars of MFI indicate the geometric standard deviation of means for three transfected cell cultures. Source data are provided as a Source Data file