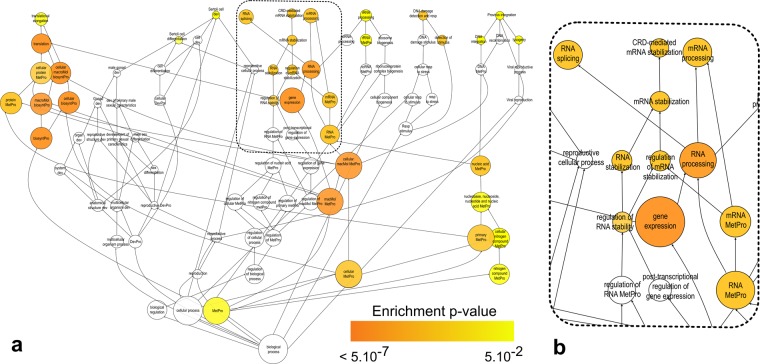
Figure 2.
Functional groups over-represented among potential M2-1 binding partners. A gene set enrichment analysis was performed using BiNGO software on all potential M2-1 binding partners. P-values were determined by a hypergeometric test (alpha = 0.01) with a Benjamini & Hochberg correction. All GO terms related to biological processes found among this selection are displayed as circles, organized from the most general at the bottom to the most specific at the top. GO terms in an inclusion relation are linked by an arrow, with the arrow’s head pointing to the daughter GO term. Each circle’ diameter varies according to the GO term’s total size, and its color varies according to its enrichment p-value. GO terms which aren’t significantly enriched in the analysis are shown as blank circles. Abbreviations used: macromolecule = macMol, metabolic process = MetPro, development = dev, biosynthetic process = BiosyntPro. Response = resp (a). Complete graph. (b) Area with GO terms related to RNA metabolism.