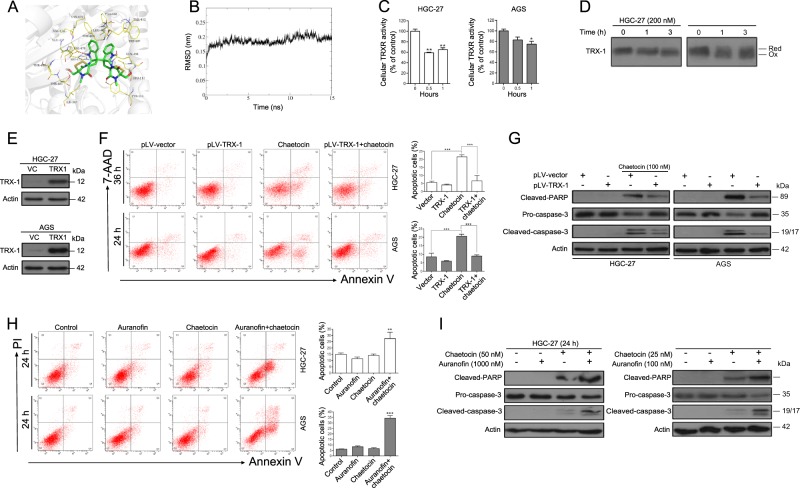
Fig. 3. Chaetocin-inactivated TRXR-1 is required for GC cell apoptosis.
a Molecular docking between chaetocin and TRXR-1 was simulated using the AutoDock 4.2 program. b RMSD trajectories of chaetocin and TRXR-1 in a 15 ns MD simulation. c TRXR-1 activity was determined by DTNB assay in HGC-27 and AGS cells treated with 200 nM and 100 nM chaetocin, respectively, for the indicated lengths of time. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, vs. control group. d Lysates of HGC-27 and AGS cells treated with 200 nM and 100 nM chaetocin, respectively, for the indicated lengths of time were alkylated by AMS and the redox state of TRX-1 was separated by non-reducing SDS-PAGE followed by western blot analysis. Reduced and oxidized forms are indicated. e TRX-1 expression levels in TRX-1-overexpressing HGC-27 and AGS cells were determined by western blot. TRX-1-overexpressing HGC-27 and AGS cells were treated with chaetocin (100 nM for HGC-27, 50 nM for AGS). f Apoptosis was analyzed by flow cytometry. Results were shown as mean ± SD of three independent experiments. ***P < 0.001. g The expression levels of cleaved-PARP and caspase-3 were detected by western blot. HGC-27 and AGS cells were cotreated with auranofin (1000 nM for HGC-27, 100 nM for AGS) and chaetocin (50 nM for HGC-27, 25 nM for AGS). h Apoptosis was analyzed by flow cytometry. Results were shown as mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001, vs. control group. i The expression levels of cleaved-PARP and caspase-3 were detected by western blot. All blots presented here are representative of three independent experiments