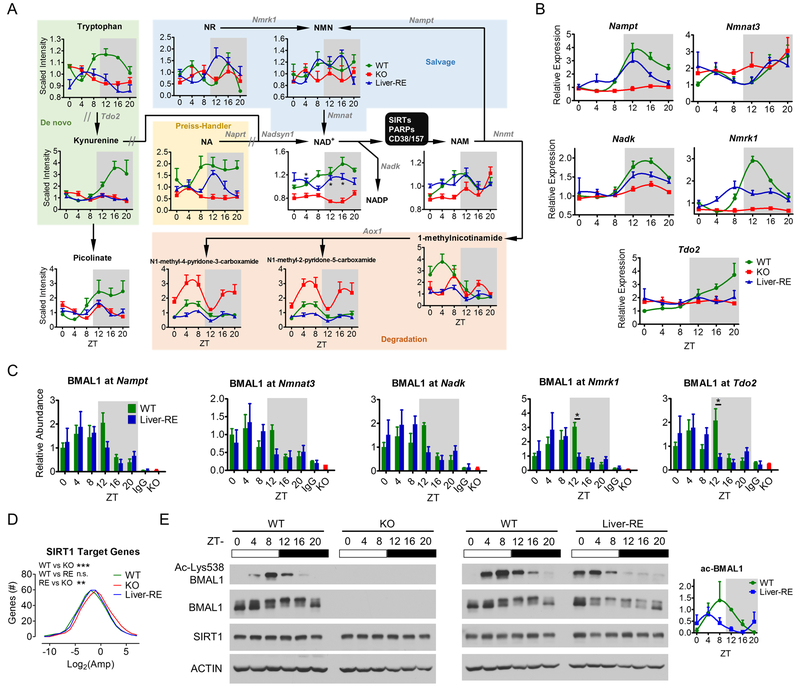
Fig. 5. Clock regulation of hepatic NAD+ metabolism.
A) Schematic of NAD+ metabolism showing the effect of organism-wide clock deficiency and reconstitution of liver clock. One-way ANOVA, *=p<0.01. Metabolite names are black: NR, nicotinamide riboside; NMN, nicotinamide mononucleotide; NA, nicotinate; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide. Enzymes are gray: Nmrk1 – nicotinamide riboside kinase 1; Nampt – nicotinamide phosphoribosyltransferase; Nmnat – nicotinamide mononucleotide adenylyltransferase; Tdo2 – tryptophan 2,3-dioxygenase; Naprt – nicotinate phosphoribosyltransferase; Nadsyn1 – glutamine-dependent NAD+ synthetase 1; Nnmt – NAM N-methyltransferase; Nadk - NAD+ kinase; Aox1 – aldehyde oxidase 1. // - multiple enzymatic steps. B) Key enzymes of NAD+ metabolism validated by qPCR. See also Fig. S4A. C) BMAL1 recruitment to NAD+-related genes, IgG and KO negative controls at ZT20. Two-way ANOVA, *=p<0.05. D) Amplitude of circadian SIRT1 target genes. One-way ANOVA, n.s.=not significant; **=p<0.01; ***=p<0.001. E) Representative western blot from liver whole-cell extracts, n=3. BMAL1 and ACTIN blots are the same as Fig. 1 and S1.