Abstract
The histone lysine demethylase 4 (Kdm4/Jmjd2/Jhdm3) family is highly conserved across species and reverses di- and tri-methylation of histone H3 lysine 9 (H3K9) and lysine 36 (H3K36) at the N-terminal tail of the core histone H3 in various metazoan species including Drosophila, C.elegans, zebrafish, mice and humans. Previous studies have shown that the Kdm4 family plays a wide variety of important biological roles in different species, including development, oncogenesis and longevity by regulating transcription, DNA damage response and apoptosis. Only two functional Kdm4 family members have been identified in Drosophila, compared to five in mammals, thus providing a simple model system. Drosophila Kdm4 loss-of-function mutants do not survive past the early 2nd instar larvae stage and display a molting defect phenotype associated with deregulated ecdysone hormone receptor signaling. To further characterize and identify additional targets of Kdm4, we employed a genome-wide approach to investigate transcriptome alterations in Kdm4 mutants versus wild-type during early development. We found evidence of increased deregulated transcripts, presumably associated with a progressive accumulation of H3K9 and H3K36 methylation through development. Gene ontology analyses found significant enrichment of terms related to the ecdysteroid hormone signaling pathway important in development, as expected, and additionally previously unidentified potential targets that warrant further investigation. Since Kdm4 is highly conserved across species, our results may be applicable more widely to other organisms and our genome-wide dataset may serve as a useful resource for further studies.
Keywords: Kdm4, Histone methylation, Drosophila, Development, Epigenetics, Histone
Introduction
The histone lysine demethylase 4 (Kdm4/Jmjd2/Jhdm3) family of histone demethylases is highly conserved across species and demonstrated to be a crucial regulator of various cellular processes. It reverses di- and tri-methylation of histone H3 lysine 9 (H3K9me2,3) and lysine 36 (H3K36me2,3) at the N-terminal tail of the core histone H3 in various metazoan species including Drosophila, C.elegans, zebrafish, mice and humans, although in Arabidopsis, its substrate was found to be histone H3 lysine 27 (H3K27) (Hillringhaus et al. 2011; Klose et al. 2006; Lu et al. 2011; Whetstine et al. 2006; Zhang et al. 2012). Various studies in different organisms have shown that Kdm4 family members play key roles in oncogenesis and development by its enzymatic activity towards H3K9me2,3 and H3K36me2,3. Furthermore, a recent study in Drosophila showed that it also catalyzes the demethylation of H3K56me3 in heterochromatin (Colmenares et al. 2017).
The Kdm4 family of demethylases was initially characterized in the context of tumorigenesis. It was found that Kdm4C is amplified in esophageal squamous cell carcinoma (Katoh and Katoh 2004) and Kdm4A overexpression was detected in various cancer cells (Gray et al. 2005). The first in vivo characterization of the biological role of Kdm4 was in C.elegans, in which its depletion in the germline resulted in increased DNA damage and apoptosis, and further studies also demonstrated its role in DNA replication by targeting H3K9me2,3 and modulating heterochromatin protein 1y (HP1γ) recruitment (Black et al. 2010, 2012; Whetstine et al. 2006). Multiple studies have linked the oncogenic potential of the Kdm4 family to its role as a co-activator of nuclear hormone receptor-mediated transcription. It was found that KDM4 interacts with the androgen receptor (AR) to mediate target gene activation by promoting the removal of the transcriptionally repressive H3K9me2,3 mark in prostate cancer cells and additionally playing a role in AR turnover (Coffey et al. 2013; Gaughan et al. 2013; Shin and Janknecht 2007; Wissmann et al. 2007). Similarly, studies in breast cancer cells found that KDM4 family members interact with the estrogen receptor (ER) and act as co-activators of target genes by removing the repressive H3K9me2,3 mark at target promoters and enhancers (Gaughan et al. 2013; Young and Hendzel 2013). Another study has described KDM4’s oncogenic role in acute myeloid leukemia, where it transcriptionally activates interleukin 3 receptor α (II3ra) and promotes survival (Agger et al. 2016). Furthermore, overexpression of various KDM4 family members resulted in defective DNA mismatch repair and genomic instability, thus suggesting another mechanism by which they contribute to tumorigenesis (Awwad and Ayoub, 2015).
Ample studies have also found that the Kdm4 family of demethylases plays key roles in stem cell differentiation and development. In murine embryonic stem cells, Kdm4C regulates self-renewal by removing H3K9me3 at the promoter of key stem cell regulator, Nanog to prevent the recruitment of transcriptionally repressive proteins, Heterochromatin Protein 1α (HP1α) and KRAB domain of KOX1 (KAP1) (Loh et al. 2007). Similarly, a different study demonstrated that conditional knockdown of Kdm4a and Kdm4c results in impaired embryonic stem cell self-renewal both in vivo and in vitro (Pedersen et al. 2016). In human mesenchymal stem cells, KDM4B regulates the transcriptional activation of Distal-less (DLX) to inhibit adipogenesis by H3K9me3 removal (Ye et al. 2012). Consistent with its ability to regulate stem cells, it also plays significant roles in organismal development. A study found that the sole rice Kdm4 gene regulates the floral organ development phenotype by demethylation of H3K9me3 at the promoters of relevant key developmental genes (Sun and Zhou 2008). Furthermore, inhibiting Kdm4A during chick embryogenesis results in downregulation of various neural crest specification genes and increased H3K9me3 enrichment at the promoter of Sox10, a key regulator in neural crest regulation (Strobl-Mazzulla et al. 2010).
In Drosophila, two Kdm4 family members have been identified and described to be functional H3K9me2,3 and H3K36me2,3 demethylases (Lloret-Llinares et al. 2008). Additional studies found that the interaction of Kdm4A with HP1a stimulates its activity towards H3K36 demethylation in vitro and that its overexpression in vivo results in male lethality with a concomitant decrease in bulk H3K36 methylation (Crona et al. 2013; Lin et al. 2008). Furthermore, Drosophila Kdm4A regulates lifespan and male-specific sex determination by transcriptional regulation of specific genes (Lorbeck et al. 2010). It has also been demonstrated that H3K9me2,3 demethylation by Drosophila Kdm4B controls the recruitment of factors involved in UV-induced DNA damage response (Palomera-Sanchez et al. 2010). A recent study described that following X-ray irradiation, Kdm4A mediates heterochromatic double-stranded DNA relocation by H3K56me3 demethylation and that Kdm4A loss-of-function, in combination with other double-stranded repair pathway mutation, impacts organismal survival (Colmenares et al. 2017). Kdm4A appears to additionally have a demethylase activity-independent role in heterochromatin organization (Colmenares et al. 2017).
Similarly to other organisms, Drosophila Kdm4 also appears to be important for development, as the loss-of-function of both Kdm4A and Kdm4B results in lethality in the early 2nd instar larval stage, with a molting defect phenotype associated with deregulation of the Ecdysone nuclear hormone receptor signaling cascade (Tsurumi et al. 2013). Given the important role of the Kdm4 family, in this study, we aimed to further elucidate additional potential transcriptional targets of Kdm4A and Kdm4B double mutants at different stages of development leading up to the time of lethality. The therapeutic potential of Kdm4 inhibitors for the treatment of various cancers (Chin and Han 2015; Duan et al. 2015; Kim et al. 2014; Lohse et al. 2011; Ye et al. 2015) and as anti-viral agents (Liang et al. 2013; Rai et al. 2010) has been demonstrated, further highlighting the relevance and importance of identifying biological mechanisms pertinent to the Kdm4 family of histone demethylases. Taking advantage of the highly conserved Drosophila system where there are only two redundant Kdm4 family members, compared to mammals where there are five, we sought to identify transcriptional targets at various stages of development comprehensively by employing a genome-wide approach. The dataset generated from this study could potentially also serve as an additional resource for further studies.
Materials and methods
Fly stocks/genetics and RNA sample preparation
All crosses were carried out at 25 °C on standard cornmeal/ agar medium. Fly stocks of Kdm4AKG04636, Kdm4BEY10737 and Sco/CyO-GFP lines were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). For creating the double mutant line, the Kdm4A and Kdm4B lines were recombined with CyO-GFP as the balancer chromosome. We outcrossed the mutants to W1118 to minimize background differences. W1118 larvae collected at comparable stages were used as the control. Eggs were laid on an apple agar plate with yeast paste and early 1st, late 1st and early 2nd instar larvae were synchronized by egg laying time and morphology. Homozygous larvae were selected based on the lack of the GFP marker. The larvae were washed twice with deionized water and total RNA was prepared using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s manual. RNA quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies Inc., Palo Alto, CA, USA).
Microarray analyses
To prepare microarray samples from the RNA isolated, 200 ng of total RNA was used to prepare biotin-labeled RNA using Ambion MessageAmp Premier RNA Amplification Kit (Applied Biosystems, Forster City, CA, USA). Briefly, the first strand of cDNA was synthesized using ArrayScript reverse transcriptase and an oligo(dT) primer bearing a T7 promoter. Then, DNA polymerase I was used (in the presence of E. coli RNase H and DNA ligase) to convert single-stranded cDNA into double-stranded DNA (dsDNA), which was then used as a template for in vitro transcription in a reaction containing biotin-labeled UTP and T7 RNA Polymerase to generate biotin-labeled antisense RNA (aRNA). Twenty |jg of labeled aRNA was fragmented and 15 μg of the fragmented aRNA was hybridized to Affymetrix Drosophila Genome 2.0 Array Chips according to the manufacturer’s manual (Affymetrix, Santa Clara, CA, USA). Array Chips were stained with streptavidin-phycoerythrin, followed by an antibody solution (anti-streptavidin) and a second streptavidin-phycoerythrin solution, performed by a GeneChip Fluidics Station 450. The Array Chips were scanned with the Affymetrix GeneChip Scanner 3000.
R version 3.1.3 was used for the subsequent analyses. GCRMA was used for the numerical conversion to expression intensity (Wu et al. 2004), using the R packages, Affy and EMA (Gautier et al. 2004; Gentleman et al. 2004; Servant et al. 2010). The end result yielded normalized log2 expression intensity of each of the probe sets for each sample (Supplementary Material 1). Control probe sets were filtered, as well as those where the wild-type and respective mutant intensities all had log2 intensity below the 2.5 threshold as determined appropriate by the histogram plot in EMA. After these filtering steps, the total number of probe sets included for subsequent analyses was 10,724. Pearson-Ward metrics were used for hierarchical clustering and heatmap plot of the top 2000 differentially expressed probe sets.
Functional assessment of down- and upregulated genes
The log2 fold change of the Kdm4 mutant over wild-type samples was determined. Further analyses of assessing stage-specific differences of mutants were conducted using a 2-fold change cutoff (Supplementary Material 3), as this cutoff corresponds to the top 15th percentile upregulated and downregulated log2 fold change of all probes, which were found to be 1.01 and – 1.09, respectively. As expected, the probe set for Kdm4A (1635774_at) showed more than the cutoff of 2-fold downregulation in the Kdm4 mutant samples at all stages investigated. The probe set for Kdm4B (1629788_at) is labeled as also targeting CG17724 that shares overlapping genomic regions and, therefore, not expected to provide a reliable quantification of Kdm4B.
Database for Annotation, Visualization and Integrated Discovery (DAVID), version 6.8 was used for functional annotation and assessing the top 20 Fold Enrichment of Gene Ontology terms of putative H3K9me3 and H3K36me3-regulated gene probe sets (Dennis et al. 2003). Classification of different analyses groups into “ecdysone-related” vs. “non ecdysone-related” loci was determined by GO terms related to the ecdysteroid hormone pathway, chitin-based processes, molting, instar larval/pupal development and metamorphosis, or by previously published results that found evidence of regulation by ecdysone signaling by a genome-wide approach using mutants or 20-hydroxy- ecdysone (20E) hormone responsiveness (Bechstead et al. 2005; Davis and Li 2013; Gauhar et al. 2009; Gonsalves et al. 2011). To test whether the “ecdysone-related” probe sets were significantly enriched for the analyses groups given the background pool of all the 10,724 probe sets used for analyses (Supplementary Material 2), a hypergeometric test was performed to calculate the p value.
Analysis of modENCODE H3K9me3 and H3K36me3 enrichment sites
In the modEncode database (modEncode Consortium et al. 2010), H3K9me3 and H3K36 Chromatin Immunoprecipitation-sequencing (ChIP-seq) data are available for Oregon-R whole organisms at developmental stage 14–16 h after egg laying (AEL) and during the 3rd instar larval stage (sample numbers 4939, 4950, 4952 and 4941). First, we determined enrichment loci that exist in the early embryonic stage that did not show enrichment in the larval stage, as putative Kdm4-demethylated sites through development. We therefore, analyzed these developmentally removed H3K9me3 and H3K36me3 sites and assessed overlaps with loci of differentially regulated transcripts in the early late 2nd instar transcriptome dataset, when we expect these differences to be most relevant.
RT-qPCR and ChIP-qPCR validation
Early 2nd instar Kdm4 double mutant or W1118 control larvae were collected as described in “Fly stocks/genetics and RNA sample preparation” section. For RT-qPCR, total RNA was isolated using RNeasy Plus Mini kit (Qiagen), as described in “Fly stocks/genetics and RNA sample preparation” section. The SuperScript III First-Strand Synthesis System (Thermo Fisher) was used to generate cDNA, according to the manufacturer’s manual, and subjected to Sybr Green qPCR. Expression values are shown as normalized values relative to rp49.
Primer sequences are as follows:
rp49 Forward: TCCTACCAGCTTCAAGATGAC.
rp49 Reverse: CACGTTGTGCACCAGGAACT.
spok Forward: CACTCGCTGCATAGTGGTAAA.
spok Reverse: CCGCCAAAGAGCTTGTGATA.
Eip71CD Forward: GGTGCTGGAAATCGACTATGA.
Eip71CD Reverse: CCTCATCGTGGTACAGAATCAA.
ImpE2 Forward: GGCGCTAGTGAACACATCTT.
ImpE2 Reverse: GAGTACTCTGGCTTGGCTAATG.
Eip78C Forward: CACCCAAGATGACCAGCTTAT.
Eip78C Reverse: CCATCGTCCAGTGTCAATGT.
scu Forward: TCGGTCGTCTGGATCTGACT.
scu Reverse: AACGTGCCCACGGTATTGAT.
forked Forward: CTTCTTTTTGCCCCGAAGGC.
forked Reverse: GAGTACTCTACGCGACACCG.
Cpr12A Forward: GATGGAACCGCTCGCTATGA.
Cpr12A Reverse: AAGACGGTGATGTAACGCCC.
For ChIP-qPCR, samples were crushed in 200ul of 1X PBS with a pestle. Formaldehyde was added to a final concentration of 1% and samples were incubated for 15 min in room temperature, then glycine was added to a final concentration of 0.125 mM and incubated for 5 min at room temperature. The sample was centrifuged at 4000g and washed 3 times with PBS-T (1X PBS, pH 7.6 with 0.3% Triton-X). Lysis Buffer (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton-X, 0.1% Sodium Deoxycholate, 5 mM PMSF, 1X PIC) was prepared and 200 μl was added to each sample. The cells were sonicated 8 times with 15 s pulses and with 1 min rest in between pulses at 4 °C, with a Branson S-450 Sonifier using a setting of 40% and output of 5. The samples were centrifuged at 13,000g for 2 min at 4 °C to remove cell debris. 1% input control was removed and the remaining chromatin lysate was incubated overnight at 4 °C in 4 μg of anti-H3K9me3 (Upstate 07–442) or IgG control. 20 μl of pre-blocked Agarose-A beads was added to each sample and incubated overnight at 4 °C. The beads were washed three times with Wash Solution (0.1% SDS, 1% Triton-X, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl pH 8.0), then in Final Wash Solution (0.1% SDS, 1% Triton-X, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-Cl, pH 8.0) for 2 h at 4 °C. The beads were then centrifuged and the wash solution was removed. Then, the beads were incubated in 100 μl Elution Buffer 1% SDS, 100 mM NaHCO3) at 65 °C overnight to de-crosslink, followed by centrifugation and the QIAquick PCR Purification Kit (Qiagen) was used to purify the DNA from the supernatant. The purified DNA was then subjected to Sybr Green qPCR. Relative enrichment of H3K9me3 of target genes was calculated and normalized to actin5C proximal promoter region enrichment.
Primer sequences are as follows:
spok promoter Forward: GCAGACAGATGGATACGG TTAG.
spok promoter Reverse: CAGCCTTAGTAAATAGTT CTCAACATAC.
Eip71CD promoter Forward: AATCGGGAGAGGGAG AAAGA.
Eip71CD promoter Reverse: TTTCTACGCGAATGT GGAGAG.
ImpE2 promoter Forward: TCGAGTCAACAAGGAATG AGAG.
ImpE2 promoter Reverse: ACCAACTGTGCAGCGATT A.
Eip78C promoter Forward: CTTGTGTGGCTGCTGTTA TTG.
Eip78C promoter Reverse: CGAGTACTGGAGGCTCTA TCT.
scu promoter Forward: TTGCCTGCTCGAGGTAATTT.
scu promoter Reverse: GGGCTCCTATCATTGGCT TAG.
forked promoter Forward: CTGCGTGGTAGAGTATTC ACAG.
forked promoter Reverse: AGCCCGAAATTATCCCAA AGA.
Cpr12A promoter Forward: AGTTAGCTGGCTTATTGC TAGG.
Cpr12A promoter Reverse: TATCCGAAAGGGTGACTG AGA.
actin5C proximal promoter Forward: ATTCAACACACC AGCGCTCTCCTT.
actin5C proximal promoter Reverse: ACCGCACGGTTT GAAAGGAATGAC.
Results
Kdm4 double mutants show progressively increased number of differentially regulated genes
We conducted hierarchical clustering with the top 2000 differentially expressed probe sets and found that in the early 1st instar larval stage, the wild-type and Kdm4 double mutants clustered together. However, later in development, in the late 1st and early 2nd stages, wild-type and Kdm4 double mutants separated out according to the genotype, suggesting that transcriptome pattern differences between the two genotypes become more apparent in the later stages of development (Fig. 1a). When comparing the number of probe sets showing 2-fold down- and upregulation compared to the wild-type during their respective larval stages, we found that the number of downregulated probe sets increased progressively and drastically from 482 in the early 1st, to 767 in the late 1st, and to 1685 in the early 2nd stage (Fig. 1b). The number of upregulated probe sets was similar between the early and late 1st instar larval stages (417 versus 413, respectively), but increased significantly in the early 2nd larval stage (1516) (Fig. 1b). For the early 1st down- and upregulated probe sets, there were more overlaps with the early 2nd instar (294 among downregulated and 165 among upregulated probe sets) compared to the late 1st instar (167 among downregulated and 83 among upregulated probe sets). This may be due to the cyclic nature of developmental signaling, in which there are common processes that are turned on in the earlier and later stages of each instar larval stage. However, in general, the most overlaps were found between the late 1st and early 2nd instar stages (502 among downregulated and 277 among upregulated probe sets). There were 128 downregulated probe sets that were shared between the three stages, whereas for the upregulated probe sets, there were only 63.
Fig. 1.
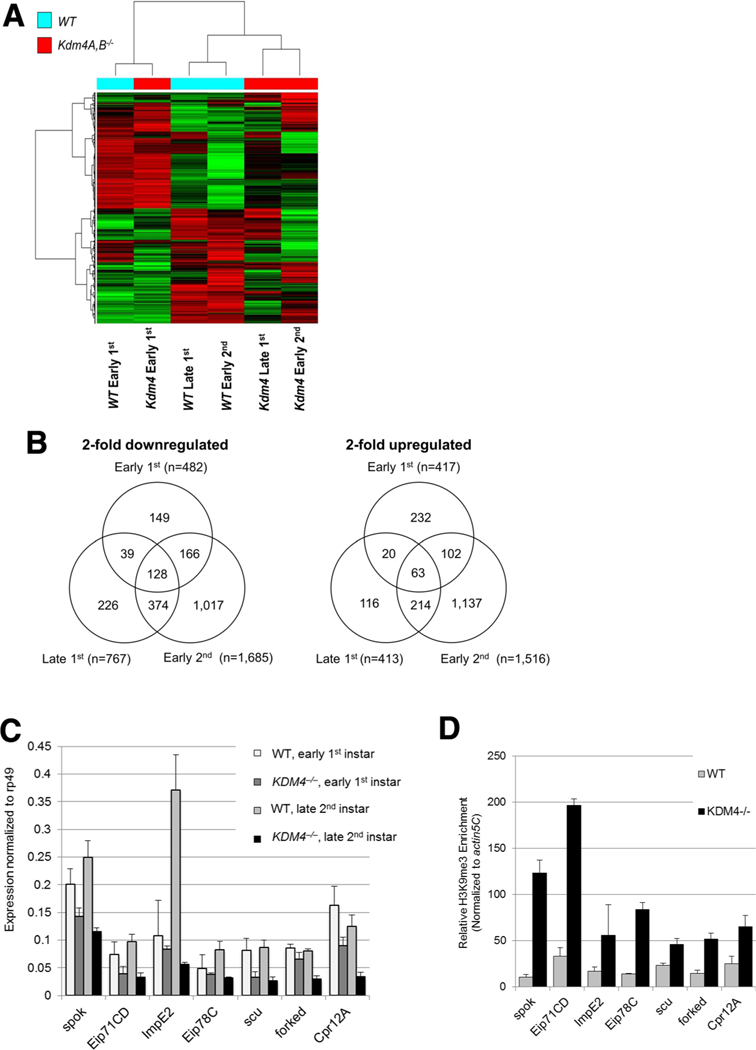
Kdm4 double mutants appear to have progressive accumulation of differentially regulated genes a Hierarchical clustering was performed using the top 2000 expressed probe sets among wild-type and Kdm4A,B double mutant larval samples at early 1st, late 1st and early 2nd instar larval stages to assess overall changes in gene expression. b The number of probe sets with above 2-fold change down- and upregula-tion compared to the wild-type was quantified and overlaps among the different stages were quantified. c RT-qPCR was performed to validate a subset of genes (spok, Eip71CD, ImpE2, Eip78C, scu, forked, Cpr12A) and found to be downregulated at least 2-fold in Kdm4A,B mutants compared to the wild-type during the 2nd instar larval stage, but not in the early 1st instar in the microarray. Expression values normalized to rp49 are plotted. d ChlP-qPCR of the downregulated gene subset was performed to assess H3K9me3 in the early 2nd instar larval stage and relative H3K9me3 enrichment normalized to actin5C proximal promoter is shown
Since the ecdysteroid pathway-related developmental phenotype is most discernable, we focused on validating the select panel of genes relevant to this process (spok, Eip71CD, ImpE2, Eip78C, scu, forked, Cpr12A) that showed downregulation in the early 2nd instar larvae in the microarray results. We verified transcript levels by RT-qPCR (Fig. 1c) and, indeed, found more than 2-fold downregulation of all these genes in the early 2nd instar Kdm4 mutants compared to the wild-type larvae, whereas in the early 1st instar stage, none was significantly more down- regulated than the 2-fold threshold. We also assessed the promoter (within 1 kb upstream region) H3K9me3 enrichment of these genes by ChIP-qPCR and detected a concomitant increase in the repressive H3K9me3 mark in the early 2nd instar larvae (Fig. 1d).
Transcriptome alternations in Kdm4 double mutants were detected for biological pathways relevant to development and other previously unlinked processes
The genome-wide approach found altered transcript levels of potential Kdm4 targets that have not previously been identified that may warrant further investigation. One such potential target is transposable elements, which were mostly found among the upregulated set, as compared to the down-regulated set at each stage. In the early 1st instar stage, 2 among 482 downregulated probe sets mapped to transposable elements, compared to 5 among 417 in the upregulated set (Fisher’s exact test p = 0.26) (Supplementary Material 3). Significant differences could be detected in the late 1st instar stage, where no transposable elements were found among the 767 downregulated probe sets, while 4 among 413 were found in the upregulated set (Fisher’s exact test p = 0.015), and further difference was seen in the early 2nd instar stage where none of the 1685 downregulated probe sets mapped to transposable elements compared to 9 among the 1516 upregulated probe sets (Fisher’s exact test p = 0.0012).
Furthermore, we conducted Gene Ontology (GO) enrichment analyses and found that among significantly altered Biological Processes terms, the most prevalent terms for all the developmental stages investigated were those relevant to ecdysteroid signaling-mediated molting behavior, as expected from previously published phenotypes (Tsurumi et al. 2013). In the early 1st instar larval stage, GO biological processes terms relevant to the hormone biosynthesis pathway, molting behavior and cuticle development appeared among the top fold enrichment and significantly altered terms (Fig. 2a; Supplementary Material 4). Similar terms related to these pathways were also found to be significant in the late 1st instar larval stage among differentially regulated probe sets (Fig. 2b; Supplementary Material 4). Comparable with the findings in early and late 1st instar larvae, in early 2nd instar larvae, significant terms also included those relevant to the ecdysteroid hormone pathway, molting behavior and cuticle development (Fig. 2c; Supplementary Material 4). GO terms pertinent to development apart from ecdysteroid signaling-mediated molting behavior included those related to wing imaginal disc development in early 1st instar larvae, morphogenesis, cell polarity and various imaginal discs in late 1st instar larvae, and in addition to these terms, dendrite morphogenesis and eye photoreceptor development were also detected in early 2nd instar (Fig. 2a–c; Supplementary Material 4). In the early 2nd instar larval stage, “determination of adult lifespan” appeared among significantly altered GO terms (Supplementary Material 4), which is also consistent with a previous publication that reported that Kdm4A mutants have a shortened lifespan phenotype (Lorbeck et al. 2010).
Fig. 2.
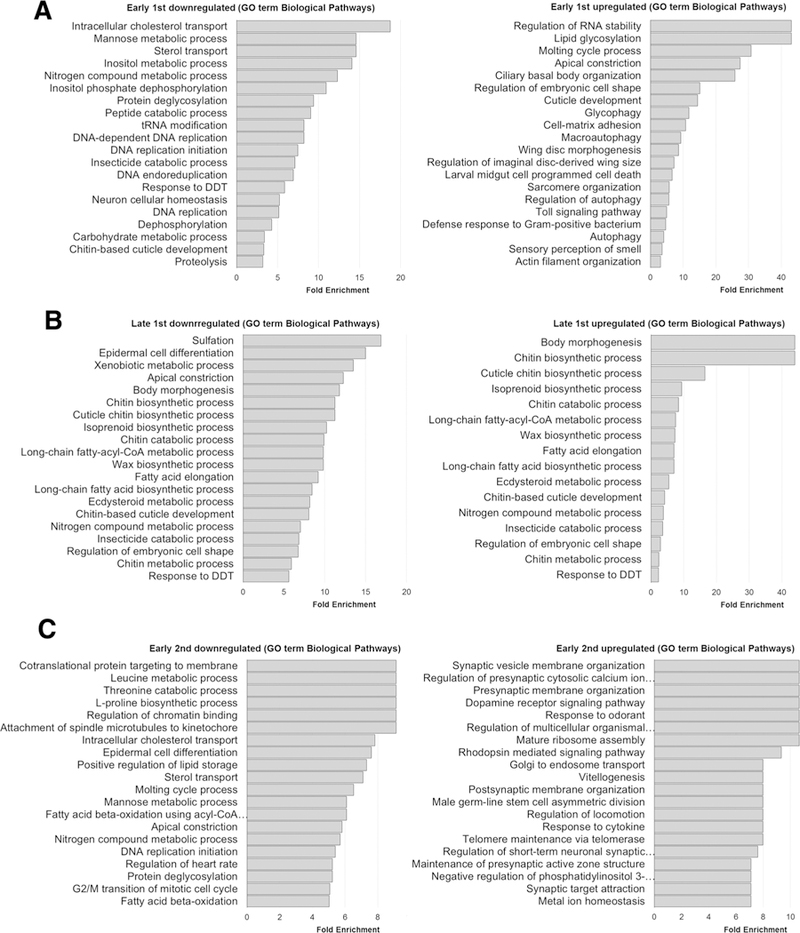
Gene ontology analyses found significant enrichment in biological processes terms relevant to ecdysone signaling and other developmental processes and signaling cascades in Kdm4 double mutants. GO enrichment analyses were performed using probe sets with at least 2-fold down- and upregulation in the a early 1st, b late 1st and c early 2nd instar larval stages, d common across the three stages, and biological process terms with top 20-fold change are shown in descending order
Various GO terms relevant to DNA replication and the cell cycle were found in both early 1st and 2nd instar downregulated sets and additionally, terms relevant to mitosis were also significantly enriched in the early 2nd instar larvae. Among early 2nd instar larvae upregulated probe sets, GO terms relevant to male germ-line maintenance, telomere maintenance, double-stranded DNA repair, apoptosis and transcription also appeared as significant terms, which have all been implicated in mechanisms related to H3K9me2,3 and H3K36me2,3-dependent transcriptional/post-transcriptional regulation and heterochromatin organization (Fig. 2a–c; Supplementary Material 4).
It may be interesting to note that terms relevant to immunity were found uniquely among upregulated genes in each of the developmental stages investigated, but not in the downregulated set. The molecular mechanism involving H3K9me2,3 in transcriptional silencing would suggest that the upregulated genes may be indirect effects. Upregulated gene sets were enriched with various GO terms relevant to the innate immune response and the Toll signaling path-way in all the three stages. GO terms pertaining to neuronal and sensory behavior, and odorant and light response were mostly found in the upregulated set in all the three developmental stages (Fig. 2a–c; Supplementary Material 4). There is also indication that signaling cascades other than ecdysteroid signaling may be impacted. GO terms identified among the early 2nd instar differentially regulated set include those for Toll and peptidoglycan recognition pathways, tyrosine receptor kinase (RTK), epidermal growth factor receptor (EGFR), phosphatidylinositol 3-kinase (PIP3K) and G-protein coupled receptor signaling pathways (Fig. 2c; Supplementary Material 4). GO terms relevant to metabolism were found frequently in both down- and upregulated sets in each of the developmental stages investigated (Fig. 2a–c; Supplementary Material 4). Further studies validating these GO term analyses would be beneficial.
We classified commonly down- and upregulated probe sets shared between the three developmental stages into “ecdysone-related” vs. “non ecdysone-related” loci by GO terms and previously published genome-wide studies (Bech-stead et al. 2005; Davis and Li 2013; Gauhar et al. 2009; Gonsalves et al. 2011). When we classified the entire pool of probe sets used for the analyses (i.e., background set), we found 38.2% (4094 out of 10,724 total) to be “ecdysone-related” (Fig. 3a; Supplementary Material 2). Among the downregulated set, 61.7% (79 probe sets out of 128) were classified to be “ecdysone-related” and among the upregulated set, 52.4% (33 probe sets out of 63) were found (Fig. 3b; Supplementary Material 3). The enrichment of “ecdysone-related” probe sets was statistically significant for both groups (p < 0.001 and p = 0.015, respectively).
Fig. 3.
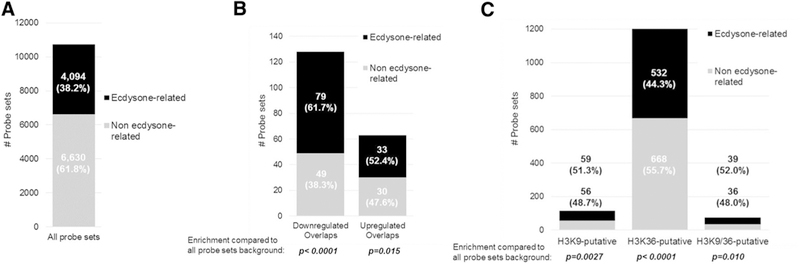
Commonly altered transcripts and putative H3K9/K36me3 developmentally altered target regions are enriched with “ecdysone-related” genes. We classified a the background pool of all probe sets prior to analyses, b those commonly altered in the early 1st, late 1st and early 2nd instar larval stages, and c those representing potential H3K9me3 and/or H3K36me3 2nd instar stage target regions into “ecdysone-related” vs. “non ecdysone-related” genes. Statistical significance of enrichment of “ecdysone-related” genes was calculated using the hypergeometric test
modENCODE ChIP-seq H3K9me3 and H3K36me3 developmentally regulated sites overlapped with various up- and downregulated genes
Taking advantage of the publicly available modENCODE database (modEncode Consortium et al. 2010) in which ChIP-seq data for H3K9me3 and H3K36me3 are available for Oregon-R early embryonic and 3rd instar larval whole animal samples, we postulated that genomic regions showing enrichment in early development that is later removed in the 3rd instar larval stage likely reflect loci that are demethylated by Kdm4 in the 2nd instar larval stage. Thus, failure of Kdm4A,B mutants to remove these marks may be one molecular mechanism that underlies the lethality phenotype. To further narrow down candidate direct targets of H3K9me3 separately from H3K36me3, we assessed overlaps between these developmentally demethylated H3K9me3 and H3K36me3 regions with corresponding altered transcripts. We evaluated downregulated probe sets for putative H3K9me3 targets and both down- and upregulated probe sets for putative H3K36me3 targets (Supplementary Material 5).
Various genes relevant to the EcR pathway, molting behavior, or cuticle formation were found among downregulated genes potentially regulated by Kdm4’s H3K9me3 activity, including several Ecdysone-inducible (Eip), cuticle and ecdysone hormone biosynthesis genes (Table 1). For putative H3K9me3-regulated genes that were downregulated in the Kdm4A,B mutants, we also found several genes that perform critical cellular functions and required for cell viability, or for which loss-of-function mutation on their own leads to developmental lethality. Thus, they may also contribute to the nonviable Kdm4A,B phenotype (Supplementary Material 6; Apger et al. 2010; Cosgrove et al. 2012; Hernández et al. 2004; Jaspers et al. 2014; LaLonde et al. 2006; Larschan et al. 2007; Leshko-Lindsay and Corces 1997; Maynard et al. 2010; Ocorr et al. 2007; Ono et al. 2006; Schittenhelm et al. 2009; Schwed et al. 2001; Shtorch et al. 1995; Technau and Roth 2008; Torroja et al. 1998; Wang et al. 2003; Zhang and Ward 2010; Zhu et al. 2008). Since H3K36me3 is known to regulate post-transcriptional alternative splicing events, we considered both down- and upregulation of different isoforms as putative direct targets (Table 2). Noteworthy examples include multiple Ecdysone-inducible (Eip) and cuticle formation genes, and other highly relevant components, such as the ecdysone hormone biosynthesis enzyme, shadow (sad), Ecdysis triggering hormone (ETH) and Hormone receptor-like in 38 (Hr38). We also assessed whether “ecdysone-related” probe sets were significantly enriched among the group representing putative H3K9me3, H3K36me3 and H3K9/ K36me3 common targets. For the putative H3K9me3 target group, the proportion of “ecdysone-related” sites was found to be 51.3% (59 out of 115, p = 0.0027); among putative H3K36me3 targets, this proportion was 44.3% (532 out of 1200, p < 0.0001) and among putative H3K9/ K36me3 common targets, it was 52.0% (39 out of 75, p = 0.010) (Fig. 3c; Supplementary Material 5).
Table 1.
Ecdysteroid pathway and molting behavior-related early 2nd instar downregulated putative H3K9me3 target transcripts
| Probe set | Gene | Transcript |
|---|---|---|
| Downregulated | ||
| 1638505_at | tan (t) | CG12120-RA |
| 1626830_at | Larval cuticle protein 1 (Lcpl) | CG11650-RA |
| 1625276_a_at Ecdysone-induced protein 28/29kD (Eip71CD) |
CG7266-RC | |
| 1632561_at | Laccase 2 (laccase2) | CG30437-RC |
| 1626695_at | CG9518 | CG9518-RA |
| 1625621_s_at forked (f) | CG5424-RB | |
| 1634433_at | Scully (scu) | CG7113-RA |
| 1622932_s_at singed (sn) | CG32858-RC | |
| 1638601_at | Spookier (spok) | CG40123-RA |
| 1634928_at | Ecdysone-inducible gene E2 (ImpE2) | CG1934-RA |
| 1623060_at | Cuticular protein 12A (Cpr12A) | CG15757-RA |
| 1640770_a_at Ecdysone-induced protein 78C (Eip78C) |
CG18023-RA | |
| 1638227_at | Uninflatable (uif) | CG9138-RA |
Table 2.
Ecdysteroid pathway and molting behavior-related early 2nd instar downregulated and upregulated putative H3K36me3 target transcripts
| Probe set | Gene | Transcript |
|---|---|---|
| Downregulated | ||
| 1623060_at | Cuticular protein 12A (Cpr12A) | CG15757-RA |
| 1623494_at | Ecdysone-inducible gene E3 (ImpE3) | CG2723-RA |
| 1623668_at | Shavenoid (sha) | CG13209-RA |
| 1624121_at | Knickkopf (knk) | CG6217-RA |
| 1625470_s_at | CG1520 (WASp) | CG1520-RA |
| 1625621_s_at | Forked (f) | CG5424-RB |
| 1626235_at | Miniature (m) | CG9369-RA |
| 1626485_at | Shadow (sad) | CG14728-RA |
| 1626841_s_at | Crinkled (ck) | CG7595-RA |
| 1626984_at | Glucose dehydrogenase (Gld) | CG1152-RA |
| 1627051_at | Larval cuticle protein 9 (Lcp9) | CG16914-RA |
| 1627571_at | Cyclic-AMP response element binding protein A (CrebA) | CG7450-RA |
| 1628067_s_at | Dusky-like (dyl) | CG15013-RB |
| 1628301_at | Doublesex cognate 73A (dsx-c73A) | CG32159-RB |
| 1628930_at | CG14485 (swi2) | CG14485-RA |
| 1628938_at | Sec24CD ortholog (Sec24CD) | CG10882-RA |
| 1631375_a_at | Ecdysone-inducible gene E1 (ImpE1) | CG32356-RB |
| 1631774_at | Cuticular protein 66Cb (Cpr66Cb) | CG7076-RA |
| 1632378_at | No mechanoreceptor potential A (nompA) | CG13207-RA |
| 1632561_at | Laccase 2 (laccase2) | CG30437-RC |
| 1633919_s_at | Lethal (3) malignant blood neoplasm (l(3)mbn) | CG12755-RA |
| 1634766_at | Serrate (Ser) | CG6127-RA |
| 1637182_at | CG9503 | CG9503-RA |
| 1638610_at | Neyo (neo) | CG7802-RA |
| 1638663_at | Sec61 alpha subunit (Sec61alpha) | CG9539-RA |
| 1639979_at | CG8927 | CG8927-RA |
| 1640770_a_at | Ecdysone-induced protein 78C (Eip78C) | CG18023-RA |
| Upregulated | ||
| 1623084_at | Shaggy (sgg) | CG2621-RA |
| 1623160_at | Matrix metalloproteinase 1 (Mmp1) | CG4859-RA |
| 1623164_a_at | Ecdysone-induced protein 75B (Eip75B) | CG8127-RB |
| 1628927_at | CG17914 gene product from transcript CG17914-RB (yellow-b) | CG17914-RA |
| 1629747_at | Cuticular protein 49Ag (Cpr49Ag) | CG8511-RA |
| 1630916_at | MAP kinase kinase 4 (Mkk4) | CG9738-RA |
| 1631481_a_at | Kruppel homolog 1 (Kr-h1) | CG18783-RA |
| 1631765_at | Puckered (puc) | CG7850-RA |
| 1634405_s_at | p21 -activated kinase (Pak) | CG10295-RC |
| 1634573_a_at | Grainy head (grh) | CG5058-RC |
| 1635128_a_at | Absent, small, or homeotic discs 2 (ash2) | CG6677-RA |
| 1635331_at | Cuticular protein 49Af (Cpr49Af) | CG8510-RA |
| 1635998_at | Cuticular protein 67B (Cpr67B) | CG3672-RA |
| 1636202_s_at | Ecdysone-inducible gene L2 (ImpL2) | CG15009-RA |
| 1636630_s_at | Garnysstan (gny) | CG5091-RA |
| 1637421_at | Chitinase 2 (Cht2) | CG2054-RA |
| 1639278_at | Ecdysis triggering hormone (ETH) | CG18105-RA |
| 1639366_at | Hormone receptor-like in 38 (Hr38) | CG1864-RC |
| 1639823_at | Ebony (e) | CG3331-RA |
Discussion
This study investigated the genome-wide transcriptional profile of mutants of the Drosophila Kdm4 family of H3K9me2,3 and H3K36me2,3 dual demethylases. Considering the highly conserved function of Kdm4, as well as its substrates H3K9me2,3 and H3K36me2,3, the findings of this study may also be applicable to furthering the understanding of their in vivo functions in other organisms. Since there are only two members of the Kdm4 family, A and B in Drosophila and the loss-of-function of both genes were found to have a drastic phenotype of lethality with molting defect associated with deregulated ecdysteroid hormone signaling cascade (Tsurumi et al. 2013), our findings are expected to be biologically relevant. Our study may provide insights into the regulatory role of Kdm4 in transcriptional activation by H3K9me2,3 demethylase activity and in post-transcriptional alternative splicing by H3K36me2,3 demethylase activity that may contribute to cellular events leading to the observed phenotypic outcomes (Fig. 4).
Fig. 4.
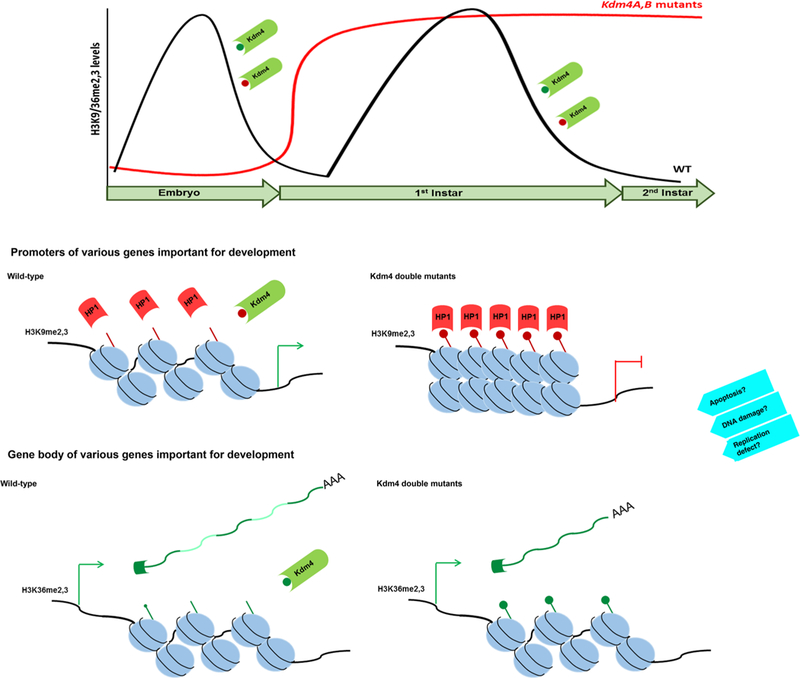
Model of the transcriptional basis of lethality in Kdm4 double mutants. In wild-type animals, Kdm4A and/or Kdm4B remove H3K9me2,3 at developmental gene loci to regulate transcriptional activation, whereas in the Kdm4A,B double mutants, H3K9me2,3 hypermethylation of direct target genes results in gene silencing by HP1a recruitment. In the gene body of target genes, in wild-type animals, Kdm4 removes H3K36me2,3 at appropriate exons to ensure correct splice site choice; however, Kdm4A,B double mutants are hypermethylated, resulting in improper and/or over-splicing events that result in mRNA without all the correct exons. Considering cellular events other than transcriptional control and alternative splicing regulated by Kdm4 that have been reported previously, it is also likely that additional mechanisms including apoptosis, DNA damage and defects in replication may also contribute to lethality
Drosophila Kdm4 has a role as a transcriptional co-activator for the ecdysteroid hormone receptor, analogously to the mammalian system involving KDM4 members as co-activators of the Androgen Receptor and Estrogen Receptor nuclear hormone signaling, thus further promoting studies using the simple Drosophila system (Gaughan et al. 2013; Shin and Janknecht 2007; Wissmann et al. 2007). Kdm4A,B double mutants are viable until the early 2nd instar larval stage, despite the ecdysone hormonal response being normally important also during the molting process to the 1st instar stage. It is well established that during Drosophila early development, maternal products are abundant and play essential roles. It may be likely that maternal effects allow the Kdm4 double mutants to undergo the first wave of hormone signaling, whereas in the later 2nd instar stage when maternal products become less abundant or no longer present, they are unable to complete the second wave of hormone signaling, contributing to lethality. Our results that found increased altered probe sets with each successive developmental stage may suggest that Kdm4A,B double mutants progressively accumulate H3K9me2,3 and H3K36me2,3 that eventually lead to transcriptional deregulation of a large number of genes, ultimately resulting in lethality. Most overlaps in deregulated probe sets were shared between the late 1st and early 2nd instar stages.
GO terms pertinent to the ecdysteroid hormone signaling and molting behavior were among those that appeared most frequently in our differential gene analysis. We also detected significant enrichment in probe sets that are relevant to this process among deregulated probe sets shared across the developmental stages. These observations are expected, given the specific phenotype associated with Kdm4A,B mutants. Our analyses also found various GO terms related to DNA damage, apoptosis and DNA replication, consistent with observed Kdm4 loss-of-function in both C.elegans and Drosophila, where previous studies have linked these phenotypes to deregulation of H3K9me2,3 and subsequent effect on HP1a/y recruitment (Black et al. 2010, 2012; Palomera-Sanchez et al. 2010; Whetstine et al. 2006). Previous studies have also shown the role of H3K36 methylation in double-stranded break DNA repair (Jha and Strahl 2014). Considering the established role of H3K9me2,3 in heterochromatin, it is also feasible that Kdm4A,B mutants are susceptible to genomic instability, thus showing transcriptional changes in genes related to DNA damage. Our study also found a GO term relevant to adult lifespan, consistent with the observation that Drosophila Kdm4A mutant males have reduced lifespan (Lorbeck et al. 2010). Drosophila Kdm4A loss-of-function, in combination with loss-of-function of various double-stranded DNA repair component, also impacts organismal survival (Colmenares et al. 2017). HP1a also regulates lifespan (Larson et al. 2012) and thus our findings may indicate that altered H3K9me2,3 by Kdm4 mutation may contribute to deregulation of genes related to lifespan. It was previously shown that HP1a and H3K9me2,3 are important for male germ-line stem cell maintenance in Drosophila (Xing and Li 2015) and analyses also found GO terms relevant in this process. Moreover, transposable elements were only found among the upregulated probe sets in the late 1st and early 2nd instar larval stages, and may reflect the dysregulation of heterochromatin maintenance as a result of Kdm4 loss. Together with the likelihood that Kdm4 mutants have altered DNA damage response, as suggested by our findings and shown in other studies, it may be one factor contributing to lethality. Future studies validating these findings will be advantageous.
The publicly available modEncode database provided the means to analyze H3K9me3 and H3K36me3 demethylated regions through the progression of the larval developmental program, where Kdm4 demethylase activity is likely to be responsible for removing these marks. When classifying probe sets commonly deregulated across developmental stages and putative H3K9me3 and H3K36me3-mediated direct targets into “ecdysone-related” versus “non ecdysone-related” genes in the 2nd instar stage, we found significant enrichment of “ecdysone-related” probe sets. The classifications were established by GO terms related to the ecdysteroid pathway and previously published genome-wide studies (Bechstead et al. 2005; Davis and Li 2013; Gauhar et al. 2009; Gonsalves et al. 2011). However, these studies investigated ecdysteroid pathway mutants during metamorphosis or treating cells with the 20E hormone and, therefore, additional validation studies are necessary. Furthermore, among this subset of downregulated and H3K9me3-relevant sites, we also detected various genes shown to have a developmentally nonviable phenotype involving other mechanisms, which may also be contributing to Kdm4A,B lethality. Taken these analyses together, our study suggests that ecdysone signaling may be a major molecular mechanism leading to lethality in Kdm4 loss-of-function mutants, although other mechanisms are also likely to be involved. Thus, we propose that further studies will be beneficial for delineating these possibilities.
Recently, it was found that Drosophila Kdm4A has an additional histone residue target, H3K56me3, relevant to double-stranded DNA repair, as well as a demethylase-independent role (Colmenares et al. 2017). Mammalian KDM4B can function as a transcriptional co-activator of AR by catalyzing the removal of H3K9me3 at target gene promoters, meanwhile also enhancing AR protein stability by masking ubiquitination sites by protein-protein interactions, demon-strating its role independently of its catalytic activity (Coffey et al. 2013). It is possible that in Drosophila, there is also a similar Kdm4-EcR interplay that depends on both its catalytic role as a transcriptional co-activator and non-catalytic role of regulating protein stability. Conducting further meticulous genetic studies with catalytic-null versus whole gene knockouts and performing biochemical assays and protein-protein interaction studies would provide further important mechanistic understanding of catalytic versus non-catalytic roles of Kdm4.
It is also likely that additionally to its demethylase-independent function, non-histone catalytic targets of Kdm4 are also relevant. A study in human cell lines found that p53-KDM4A complex formation mediated by F-box22 (Fbxo22) promotes p53 degradation in a KDM4A demethylase activity-dependent manner, while ectopic expression of KDM4A catalytic mutant enhances p53’s interaction with PHD Finger Protein 20 (PHF20) and leads to its stabilization in a demethylase-independent manner (Johmura et al. 2016). Studies have previously found Drosophila Kdm4A mutants to be susceptible to DNA damage due to its catalytic and non-catalytic functions (Palomera-Sanchez et al. 2010; Colmenares et al. 2017). Similarly, non-catalytic mechanisms may be relevant to our study on developmental lethality.
Various non-histone catalytic targets of JmjC-domain demethylases other than KDM4 have been found in mammals, including p53, DNA methyltransferase 1 (DNMT1), signal transducer and activator of transcription 3 (STAT3), myosin phosphatase target subunit 1 (MYPT1), and NFkB (reviewed in Zhang et al. 2012). G9a and EHMT1 H3K9me2 methyltransferases and the SETDB1 H3K9me3 methyltrans-ferase add methyl marks on various signaling proteins such as p53, and epigenetic regulators including G9a itself and C/EBPp (reviewed in Zhang et al. 2012). H3K36me2 methyltransferases, Nuclear Receptor Binding SET Domain Protein 1 (NSD1) and SET And MYND Domain Containing 2 (SMYD2) also have signaling protein targets including NFkB, p53 and Retinoblastoma protein (RB) (reviewed in Zhang et al. 2012). Taken together, it is feasible to postulate that Kdm4 may also have an array of non-histone protein targets that may result in indirectly regulating transcriptional activity and post-transcriptional events, protein stability and protein-protein complex formation.
Our genome-wide approach uncovered previously unexplored putative Kdm4 transcriptional targets that would be of interest to further validate. GO terms pertinent to neural and sensory behavior found in this study may represent a novel link between Kdm4 and brain development. Various major signaling pathway cascades other than ecdysteroid signaling implicated from GO enrichment analyses include Toll, TOR, RTK, PGRP, EGFR, PIP3K and G-protein coupled receptor signaling pathways. Consistent with these signaling pathway terms, multiple GO terms related to immunity, cell growth and apoptosis were also suggested from our analyses. It is interesting to note that terms related to immunity were found among the upregulated set, suggesting a hyper-inflammatory state and that it may be due to indirect transcriptional de-regulation by Kdm4 loss-of-function. Metabolism is also a previously unidentified Kdm4 putative target, as suggested by our results. Since an established link exists between metabolism and heterochromatin (Bitterman et al. 2003; Meister et al. 2011), which Kdm4 regulates, it may indicate that Kdm4A,B mutants undergo altered metabolic states. Further investigation of these novel findings may aid in uncovering additional mechanisms relevant to Kdm4.
Since our aim was to elucidate putative Kdm4 targets in transcriptional/post-transcriptional events, we focused on H3K9me3 and H3K36me3 that previous literature suggests are most relevant. Taking into consideration the known molecular mechanisms of H3K9me2,3 and H3K36me2,3 in transcriptional and post-transcriptional processes, it is likely that heterochromatin-induced silencing by elevated levels of H3K9me2,3 would result in downregulation of direct target genes. As opposed to H3K9me2,3 which plays a role in transcriptional silencing, H3K36me2,3 regulates alternative splicing (Sorenson et al. 2016). Therefore, direct targets of Kdm4 with increased H3K36me2,3 may lead to altered expression of isoforms and reflected in both down- and upregulated transcripts. A direct approach to distinguishing between direct and indirect transcriptional/post-transcriptional targets genome-wide would be to conduct ChIP-seq analyses in the Kdm4 mutants to identify regions with increased H3K9me2,3 levels and comparing to H3K36me2,3 enrichment. Alternatively, it would also be highly informative to conduct ChIP-seq of Kdm4A and Kdm4B pull-down, although limitations in available antibodies for wild-type protein pull-down, and artificial effects with transgenic over-expression of tagged Kdm4 present challenges. Furthermore, assessing Kdm4 non-histone targets or interacting proteins could provide additional insights into molecular mechanisms. Such further studies have immense potential to elucidate distinct H3K9 versus H3K36-specifc roles of Kdm4.
Supplementary Material
Acknowledegements
This work was supported by an NIH Grant (R01CA131326) to WXL. AT was supported by the Shriners Hospitals Research Fellowship #84293. We would like to acknowledge Laura Goodfield, Yashoda Dhole, Paris Karniadakis, Jacqueline Baba and Ved Dhole for their input.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00438-019-01561-z) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare that there is no conflict of interest.
References
- Agger K, Miyagi S, Pedersen MT, Kooistra SM, Johansen JV, Helin K (2016) Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells. Genes Dev 30:1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apger J, Reubens M, Henderson L, Gouge CA, Ilic N, Zhou HH, Christensen TW (2010) Multiple functions for Drosophila mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics 185:1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awwad SW, Ayoub N (2015) Overexpression of KDM4 lysine demethylases disrupts the integrity of the DNA mismatch repair pathway. Biol Open. 4:498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstead RB, Lam G, Thummel CS (2005) The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol 6:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA (2003) Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev 67:376–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, Rinehart C, Quiton J, Walsh R, Smallwood A, Dyson NJ, Whetstine JR (2010) Conserved antagonism between JMJD2A/ KDM4A and HP1gamma during cell cycle progression. Mol Cell 40:736–748 [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48:491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin YW, Han SY (2015) KDM4 histone demethylase inhibitors for anti-cancer agents: a patent review. Expert Opin Ther Pat 25:135–144 [DOI] [PubMed] [Google Scholar]
- Coffey K, Rogerson L, Ryan-Munden C, Alkharaif D, Stockley J, Heer R, Sahadevan K, O’Neill D, Jones D, Darby S, Staller P, Mantilla A, Gaughan L, Robson CN (2013) The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res 41:4433–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Swenson JM, Langley SA, Kennedy C, Costes SV, Karpen GH (2017) Drosophila histone demethylase KDM4A has enzymatic and non-enzymatic roles in controlling heterochromatin integrity. Dev Cell 42:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove M, Ding Y, Rennie WA, Lane M, Hanes SD (2012) The Bin3 RNA methyltransferase (MePCE) targets 7SK RNA to control transcription and translation. Wiley Interdiscip Rev RNA 3:633–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crona F, Dahlberg O, Lundberg LE, Larsson J, Mannervik M (2013) Gene regulation by the lysine demethylase KDM4A in Drosophila. Dev Biol. 373:453–463 [DOI] [PubMed] [Google Scholar]
- Davis MB, Li T (2013) Genomic analysis of the ecdysone steroid signal at metamorphosis onset using ecdysoneless and EcRnull Dros-ophila melanogaster mutants. Genes Genomics. 35:21–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3. [PubMed] [Google Scholar]
- Duan L, Rai G, Roggero C, Zhang QJ, Wei Q, Ma SH, Zhou Y, Santoyo J, Martinez ED, Xiao G, Raj GV, Jadhav A, Simeonov A, Maloney DJ, Rizo J, Hsieh JT, Liu ZP (2015) KDM4/JMJD2 histone demethylase inhibitors block prostate tumor growth by suppressing the expression of AR and BMYB-regulated genes. Chem Biol 22:1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan L, Stockley J, Coffey K, O’Neill D, Jones DL, Wade M, Wright J, Moore M, Tse S, Rogerson L, Robson CN (2013) KDM4B is a master regulator of the estrogen receptor signalling cascade. Nucleic Acids Res 41:6892–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauhar Z, Sun LV, Hua S, Mason CE, Fuchs F, Li TR, Boutros M, White KP (2009) Genomic mapping of binding regions for the Ecdysone receptor protein complex. Cold Spring Harb Lab Press. 19:1006–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Moller M, Friis-Hansen L, Knudsen S (2004) Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinform 5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves SE, Neal SJ, Kehoe AS, Westwood JT (2011) Genome-wide examination of the transcriptional response to ecdysteroids 20-hydroxyecdysone and ponasterone A in Drosophila melanogaster. BMC Genomics. 12:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW, Kokkotou E, Dangond F (2005) Functional characterization of JMJD2A, a histone dea-cetylase- and retinoblastoma-binding protein. J Biol Chem 280:28507–28518 [DOI] [PubMed] [Google Scholar]
- Hernández G, Vázquez-Pianzola P, Zurbriggen A, Altmann M, Sierra J, Rivera-pomar (2004) Two functionally redundant isoforms of Drosophila melanogaster eukaryotic initiation factor 4B are involved in cap-dependent translation, cell survival, and proliferation. FEBS 271:2923–2936 [DOI] [PubMed] [Google Scholar]
- Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U (2011) Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem 286:41616–41625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers MHJ, Pflanz R, Riedel D, Kawelke S, Feussner I, Schuh R (2014) The fatty acyl-CoA reductase waterproof mediates airway clearence in Drosophila. Dev Biol 385:23–31 [DOI] [PubMed] [Google Scholar]
- Jha DK, Strahl BD (2014) An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun. 5:3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y, Sun J, Kitagawa K, Nakanishi K, Kuno T, Naiki-Ito A, Sawada Y, Miyamoto T, Okabe A, Aburatani H, Li SF, Miyoshi Takahashi S, Kitagawa M, Nakanishi M (2016) SCFFbxo22-KDM4A targets methylated p53 for degradation and regulates senescence. Nature Comm. 7:10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M (2004) Identification and characterization of JMJD2 family genes in silico. Int J Oncol 24:1623–1628 [PubMed] [Google Scholar]
- Kim TD, Fuchs JR, Schwartz E, Abdelhamid D, Etter J, Berry WL, Li C, Ihnat MA, Li PK, Janknecht R (2014) Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon cancer cells and identification of curcuminoids as JMJD2 inhibitors. Am J Transl Res. 6:236–247 [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312–316 [DOI] [PubMed] [Google Scholar]
- LaLonde M, Janssens H, Yun S, Crosby J, Redina O, Olive V, Altshuller YM, Choi S-Y, Du G, Gergen JP, Frohman MA (2006) A role for Phospholipase D in Drosopihla embryonic cellularization. BMC Dev Biol 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda M (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28:121–133 [DOI] [PubMed] [Google Scholar]
- Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX (2012) Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 8:e1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshko-Lindsay L, Corces VG (1997) The role of selectins in Drosophila eye and bristle development. Development. 124:169–180 [DOI] [PubMed] [Google Scholar]
- Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM (2013) Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL (2008) Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 32:696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F (2008) Characterization of Drosophila melanogaster JmjC + N histone demethylases. Nucleic Acids Res 36:2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21:2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse B, Kristensen JL, Kristensen LH, Agger K, Helin K, Gajhede M, Clausen RP (2011) Inhibitors of histone demethylases. Bioorg Med Chem 19:3625–3636 [DOI] [PubMed] [Google Scholar]
- Lorbeck MT, Singh N, Zervos A, Dhatta M, Lapchenko M, Yang C, Elefant F (2010) The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene 450:8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43:715–719 [DOI] [PubMed] [Google Scholar]
- Maynard JC, Pham T, Zheng T, Jockheck-Clark A, Rankin HB, Negward C, Spana EP, Nicchitta CV (2010) Gp93, the Drosophila GRP94 ortholog, is required for gut epithelial homeostasis and nutrient assimilation-coupled growth control. Dev Biol 339:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Schott S, Bedet C, Xiao Y, Rohner S, Bodennec S, Hudry B, Molin L, Solari F, Gasser SM, Palladino F (2011) Caenorhabditis elegans Heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol 12:R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modEncode Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330:1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen H-SV, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, Bodmer R (2007) KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Genetics 104:3943–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H, Rewitz KF, Shinoda T, Itoyama K, Petryk A, Rybczynoski R, Jarcho M, Warren JT, Marqués G, Shimell MJ, Gilbert LI, O’Connor MB (2006) Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol 298:550–570 [DOI] [PubMed] [Google Scholar]
- Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Rey-naud E, Zurita M (2010) Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem 285:31370–31379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MT, Kooistra SM, Radzisheuskaya A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K, Helin K (2016) Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J 35:1550–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai G, Kawamura A, Tumber A, Liang Y, Vogel JL, Arbuckle JH, Rose NR, Dexheimer TS, Foley TL, King ON, Quinn A, Mott BT, Schofield CJ, Oppermann U, Jadhav A, Simeonov A, Kristie TM, Maloney DJ (2010) Discovery of ML324, a JMJD2 demethylase inhibitor with demonstrated antiviral activity. Probe Reports from the NIH Molecular Libraries Program, Bethesda (MD) [PubMed] [Google Scholar]
- Schittenhelm RB, Chaleckis R, Lehner C (2009) Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J 28:2374–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwed G, May N, Pechersky Y, Calvi BR (2001) Drosophila mini-chromosome maintenance 6 is required for chorion gene amplify-cation and genomic replication. MBoC. 13:607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Gravier E, Gestraud P, Laurent C, Paccard C, Biton A, Brito I, Mandel J, Asselain B, Barillot E, Hupe P (2010) EMA—a R package for easy microarray data analysis. BMC Res Notes. 3:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Janknecht R (2007) Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun 359:742–746 [DOI] [PubMed] [Google Scholar]
- Shtorch A, Werczberger R, Segal D (1995) Genetic and molecular studies of apterous: a gene implicated in the juvenile hormone system Drosophila. Arch Insect Biochem Physiol 30:195–209 [DOI] [PubMed] [Google Scholar]
- Sorenson MR, Jha DK, Ucles SA, Flood DM, Strahl BD, Stevens SW, Kress TL (2016) Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol 13:412–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M (2010) Histone demethylase JmjD2A regulates neural crest specification. Dev Cell 19:460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105:13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M, Roth S (2008) The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly 2:82–91 [DOI] [PubMed] [Google Scholar]
- Torroja L, Ortuno-Sahagún D, Ferrús A, Hämmerle B, Barbas JA (1998) scully, an essential gene of Drosophila, is homologous to mammalian mitochondrial type II l-β-hydroxyacyl-CoA dehydrogenase/amyloid-ß peptide-binding protein. J Cell Biol 141:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi A, Dutta P, Shang R, Yan SJ, Li WX (2013) Drosophila Kdm4 demethylases in histone H3 lysine 9 demethylation and ecdyster-oid signaling. Sci Rep. 3:2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Sullivan KMC, Beckingham K (2003) Drosophila calmodulin mutants with specific defects in the musculature or in the nervous system. Genetics 165:1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467–481 [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R (2007) Cooperative demethylation by JMJD2C and LSD1 pro-motes androgen receptor-dependent gene expression. Nat Cell Biol 9:347–353 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman RC, Martinez-Murillo F, Spencer F (2004) A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. J Am Stat Assoc 99:909–917 [Google Scholar]
- Xing Y, Li WX (2015) Heterochromatin components in germline stem cell maintenance. Sci Rep 5:17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH, Wang CY (2012) Histone demethylases KDM4B and KDM6B pro-motes osteogenic differentiation of human MSCs. Cell Stem Cell 11:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Holowatyj A, Wu J, Liu H, Zhang L, Suzuki T, Yang ZQ (2015) Genetic alterations of KDM4 subfamily and therapeutic effect of novel demethylase inhibitor in breast cancer. Am J Cancer Res. 5:1519–1530 [PMC free article] [PubMed] [Google Scholar]
- Young LC, Hendzel MJ (2013) The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Biochem Cell Biol 91:369–377 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ward RE IV (2010) uninflatable encodes a novel ectodermal apical surface protein required for tracheal inflation in Drosophila. Dev Biol 15:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wen H, Shi X (2012) Lysine methylation: beyond histones. Acta Biochim Biophys Sin 44:14–27 [DOI] [PubMed] [Google Scholar]
- Zhu B, Pennack JA, McQuilton P, Forero MG, Mizuguchi K, Sutcliffe B, Gu C-J, Fenton JC, Hidalgo A (2008) Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol 6:e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


