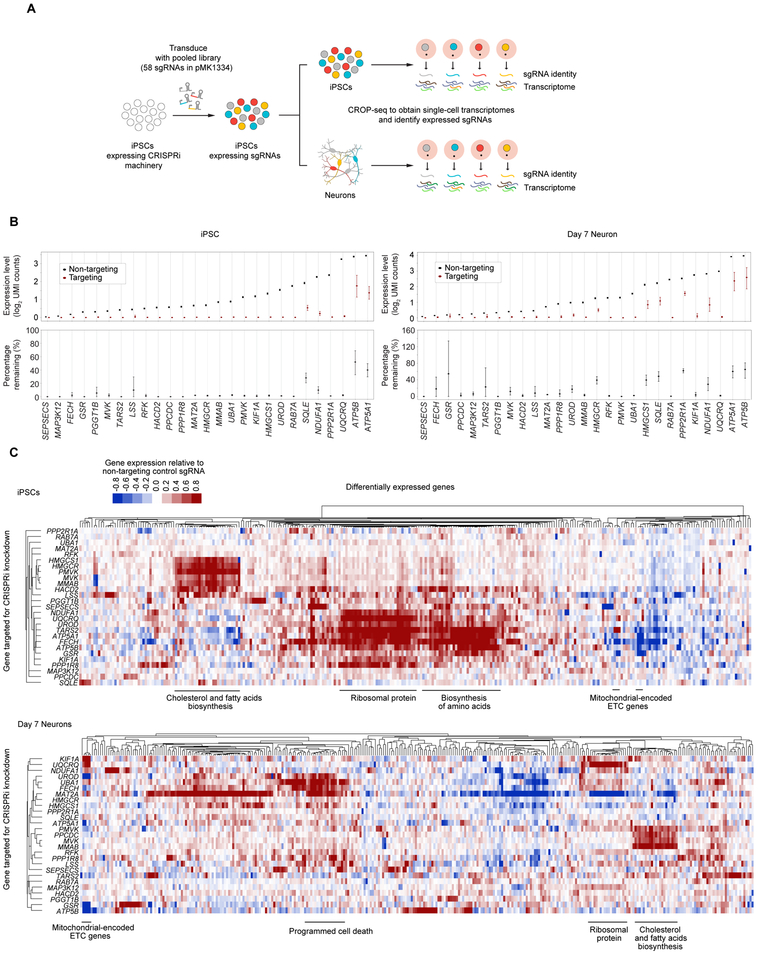
Figure 4. CROP-Seq reveals transcriptome changes in iPSCs and iPSC-derived neurons induced by knockdown of survival-relevant genes.
(A) Strategy for CROP-Seq experiments.
(B) On-target knockdown efficiencies in the CROP-Seq screen were quantified for iPSCs (left) and Day 7 neurons (right). For each target gene, the 50% of cells with the strongest on-target knockdown were selected from all cells expressing sgRNAs targeting the gene; average expression of each target gene within these cells is compared to cells with non-targeting control sgRNAs. Error bars: 95% confidence intervals estimated by bootstrapping.
(C) Changes in gene expression in response to CRISPRi knockdown of genes of interest in iPSCs (top) and Day 7 neurons (bottom). Each row represents one targeted gene; for each targeted gene, the top 20 genes with the most significantly altered expression were selected, and the merged set of these genes is represented by the columns. Rows and columns were clustered hierarchically based on Pearson correlation. Functionally related groups of differentially expressed genes are labeled.