Abstract
The mycotoxin zearalenone (ZEN) is produced by many plant pathogenic Fusarium species. It is well known for its estrogenic activity in humans and animals, but whether ZEN has a role in plant–pathogen interaction and which process it is targeting in planta was so far unclear. We found that treatment of Arabidopsis thaliana seedlings with ZEN induced transcription of the AtHSP90.1 gene. This heat shock protein (HSP) plays an important role in plant–pathogen interaction, assisting in stability and functionality of various disease resistance gene products. Inhibition of HSP90 ATPase activity impairs functionality. Because HSP90 inhibitors are known to induce HSP90 gene expression and due to the structural similarity with the known HSP90 inhibitor radicicol (RAD), we tested whether ZEN and its phase I metabolites α- and ß-zearalenol are also HSP90 ATPase inhibitors. Indeed, AtHSP90.1 and wheat TaHSP90-2 were inhibited by ZEN and ß-zearalenol, while α-zearalenol was almost inactive. Plants can efficiently glycosylate ZEN and α/ß-zearalenol. We therefore tested whether glucosylation has an effect on the inhibitory activity of these metabolites. Expression of the A. thaliana glucosyltransferase UGT73C6 conferred RAD resistance to a sensitive yeast strain. Glucosylation of RAD, ZEN, and α/ß-zearalenol abolished the in vitro inhibitory activity with recombinant HSP90 purified from Escherichia coli. In conclusion, the mycotoxin ZEN has a very prominent target in plants, HSP90, but it can be inactivated by glycosylation. This may explain why there is little evidence for a virulence function of ZEN in host plants.
Keywords: Arabidopsis, HSP90, wheat, glycosylation, Fusarium, radicicol
Introduction
The mycotoxin zearalenone (ZEN) has potent estrogenic activity in humans and animals (Kuiper-Goodman et al., 1987; Zinedine et al., 2007), and based on risk assessments, it is a regulated mycotoxin in food in Europe (European Commission, 2006b; Gromadzka et al., 2008; EFSA Panel on Contaminants in the Food Chain (CONTAM), 2011). In addition, guidance levels for feed exist which reflect differences in metabolization of ZEN in different animal species (European Commission, 2006a; European Commission, 2016). ZEN is produced by multiple species of Fusarium, particularly by Fusarium graminearum in the broad sense, which has been split into 16 species based on molecular phylogeny (Sarver et al., 2011; van der Lee et al., 2015). Also Fusarium culmorum, Fusarium crookwellense (synonym Fusarium cerealis), Fusarium equiseti, Fusarium pseudograminearum, and Fusarium semitectum consistently produce ZEN (Desjardins, 2006), as do more recently described species, such as Fusarium praegraminearum (Grafenhan et al., 2016) and Fusarium dactylidis (Aoki et al., 2015). Sporadic production has also been reported for other species (e.g., isolates of Fusarium solani (Richardson et al., 1985), Fusarium sporotrichioides (Molto et al., 1997), Fusarium oxysporum (Milano and Lopez, 1991), Fusarium tricinctum (Caldwell et al., 1970), and Fusarium heterosporum (Bottalico et al., 1989)), but this may be at least partly due to errors in identification solely based on microscopy. Nevertheless, ZEN production is widespread, particularly in pathogens of small grain cereals and corn, which suggests it might have a role as a virulence factor. Estrogen receptor signaling is absent in plants, and the biological mode of action of ZEN in planta remained so far unclear. Kim et al. (2005) reported unchanged virulence of ZEN-deficient knockout mutants on infected barley heads. Also, Gaffoor and Trail (2006) reported unaltered virulence after point inoculation of wheat heads. Likewise, Lysoe et al. (2006) observed no difference in a root rot assay with barley seedlings dipped into spore suspensions. However, gene expression studies revealed that the ZEN biosynthesis genes were not expressed under the conditions used to infect wheat and barley (Lysoe et al., 2011), and similar results have been obtained for corn (Harris et al., 2016). Concluding that ZEN plays no role in virulence might therefore be premature.
The structure of ZEN ( Figure 1 ) is similar to that of radicicol (RAD); both are members of the class of resorcylic acid lactones (Mirrington et al., 1965; Metzler, 2011). RAD (also known as monorden) was originally isolated from Nectria radicicola. RAD strongly affects expression and inhibits the activity of heat shock proteins (HSPs), particularly of the HSP90 family (Schulte et al., 1998; Roe et al., 1999; Yamada et al., 2007). Products of the HSP90 gene family play a crucial role in cellular processes such as protein folding, maturation, activation, transport, and degradation. Although most cellular proteins do not require HSP90 for folding under normal conditions, important client proteins such as transcription factors or signal transduction components (Picard, 2002) do, and consequently, HSP90 activity is essential for life.
Figure 1.
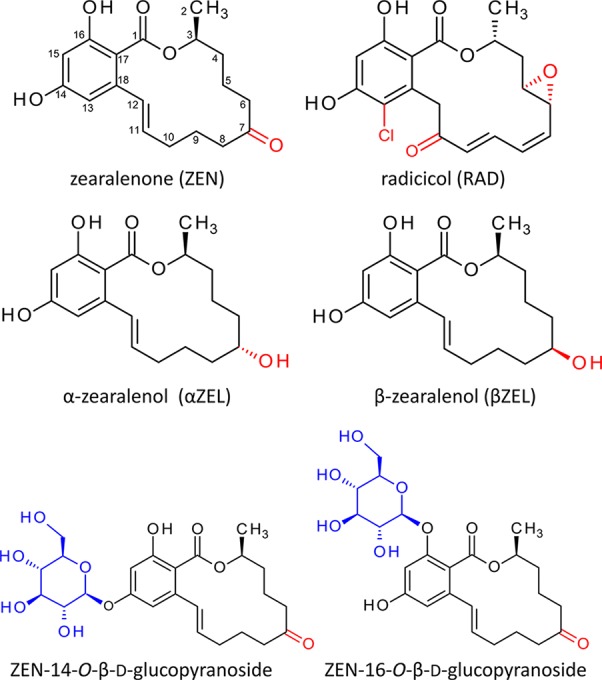
Structures of radicicol, zearalenone (Metzler, 2011), and the zearalenone phase I metabolites α- and ß-zearalenol. Structural differences are highlighted red. Below the structures of zearalenone-14-O-ß-D-glucopyranoside and zearalenone-16-O-ß-D-glucopyranoside. The glucosides of α-zearalenol and ß-zearalenol produced in this study carry the glucose moiety at the analogous position.
Binding and subsequent hydrolysis of ATP is the key mechanism through which HSP90 members interact with their client proteins. Inhibition of HSP90 ATPase activity has been thoroughly studied, and the nucleotide (ATP) binding domain (histidine kinase like ATPase superfamily, pfam02518) is highly conserved within the HSP90 family. Therefore, drugs intended to reduce HSP90 functionality, for example, in cancer therapy (Roe et al., 1999; Sgobba and Rastelli, 2009), mainly target the ATP binding site. It has been shown that RAD and the Streptomyces metabolite geldanamycin target the same N-terminal ATP binding site and strongly inhibit the ATPase activity of HSP90 members (Roe et al., 1999; Rowlands et al., 2004).
Based on results of Arabidopsis thaliana microarray data after ZEN treatment and the structural similarity between ZEN and RAD, we hypothesized that ZEN may also be an HSP90 inhibitor. In planta, the ZEN phase I plant metabolites α- and β-zearalenol (α/βZEL) and phase II glucoconjugates (Berthiller et al., 2006) are formed ( Figure 1 ). The 14-O-β-D-glucopyranosides of ZEN (ZEN14G) and α/βZEL have previously been described to occur in different plant species (Berthiller et al., 2006; Berthiller et al., 2009a; Berthiller et al., 2009b). More recently, ZEN-16-O-β-D-glucopyranoside (ZEN16G) ( Figure 1 ) has been found in ZEN-treated barley, wheat, and Brachypodium (Kovalsky Paris et al., 2014) but also in naturally infected grain samples (Nathanail et al., 2015).
Here, we tested whether ZEN and its phase I detoxification derivatives (α/βZEL) are HSP90 ATPase inhibitors and, furthermore, whether glycosylation has an effect on the HSP90 ATPase inhibitor activity.
Materials and Methods
Chemicals and Materials
In the phosphate assay, absorbance at 620 nm was determined using a PerkinElmer EnSpire 2300 Multilabel Plate Reader. F-bottom 96-well cell culture plates were obtained from Greiner Bio-One (Kremsmuenster, Austria). Malachite green, polyvinyl alcohol, ammonium molybdate, ATP sodium salt, RAD, α- and ßZEL, as well as Murashige and Skoog basal medium were purchased from Sigma-Aldrich (Vienna, Austria). In order to avoid contamination with inorganic phosphate, glassware and pH meter electrodes were rinsed extensively with double-distilled water before use.
ZEN14G, the 14-O-glucosides of α- and ßZEL, and ZEN16G were synthesized as previously reported (Poppenberger et al., 2006; Krenn et al., 2007; Berthiller et al., 2009a; Kovalsky Paris et al., 2014).
Analogously, the 16-O-glucosides of α- and ßZEL have been prepared by chemical reduction of ZEN16G with sodium borohydrate and subsequent high-performance liquid chromatography (HPLC) preparation of the mixture of αZEL-16-O-glucoside and ßZEL-16-O-glucoside (Michlmayr et al., 2017). Enhanced product ion (EPI) tandem mass spectrometry (MS/MS) scans to confirm the identity of all used glucosides were acquired on a 4000 QTrap mass spectrometer (Sciex, Framingham, USA), coupled to a 1290 UHPLC system (Agilent, Waldbronn, Germany) after negative electrospray ionization. Chromatograms and MS/MS spectra for all used ZEN metabolites can be found in the supplementary material ( Supplementary Figures S1–S6 ).
Preparation of Arabidopsis thaliana Seedlings
A. thaliana (ecotype Columbia, Col-0) seeds were surface-sterilized using sodium hypochlorite (2%) plus 0.01% Triton X for 10 min and rinsed twice with sterile water. Seeds were distributed in 24-well plates (4 wells per treatment, 30–40 seeds per well) and re-suspended in 1 ml (per well) of liquid Murashige and Skoog medium (“MS” basal salt mixture, Sigma) supplemented with 1% sucrose. After 2 days of stratification at 4°C, the plates were transferred to a growth chamber at 21°C under a 16 h light/8 h dark photoperiod.
Microarray Data
The response of the A. thaliana transcriptome to treatment with 50 µM ZEN after 2 and 24 h had been previously determined (Werner, 2005) using ATH1 22K Affymetrix gene chips at the NASC’s International Affymetrix Service (http://affymetrix.arabidopsis.info/), and the results and experimental conditions were deposited in the NASC International Affymetrix Service database as NASCARRAYS-71 and the EMBL-EBI ArrayExpress database as E-NASC-52 (http://www.ebi.ac.uk/arrayexpress/experiments/E-NASC-52/). Treatments in this experiment were with 50 µM ZEN for 2 and 24 h. As control, seedlings were treated with the same concentration of solvent (dimethyl sulfoxide, DMSO).
Expression Analysis of ATHSP90.1 by Quantitative Real-Time PCR
Total RNA was extracted from 13-day-old A. thaliana (Col-0) seedlings grown in liquid MS medium supplemented with 2.5% sucrose and 50 μM ZEN for 2 h. Samples were snap-frozen and stored at −80°C. RNA was extracted with TRI reagent TR (Molecular Research Center, Inc. Cincinnati, Ohio, USA). As control, seedlings were treated with the same concentration of solvent (DMSO). RNA was quantified with the Qubit (Invitrogen) and NanoDrop (Peqlab) systems. First-strand cDNA was synthesized from 2.5 μg RNA after RNAse free DNase I digestion (Fermentas) with Superscript III reverse transcriptase (Invitrogen) as described in Karsai et al. (2002). RT-qPCR expression analysis was performed using the Hot FirePol EvaGreen qPCR Mastermix (Solis Biodyne) with a Rotorgene 3000 (Qiagen). Primers for AtHSP90.1 were HSP90.1-RTfw 5’-ctctcacgagtgggaactcatc-3’ and AtHSP90-1-A 5’-TTGAATTCTAGCTGACCCTCC-3’ and the reference gene ADAPTOR PROTEIN-2 MU-ADAPTIN (AP2M; AT5G46630) AP2M_F 5’-CAATCGATTGCTTGGTTTGGA-3’ and AP2M_R 5’-CGAACTCGCAGACCAGATGC-3’. Absolute and relative expression were calculated with a dilution series of purified AtHSP90.1 and AP2M PCR fragments of known molar concentration in each RT-qPCR run. The amplicon sizes of AtHSP90.1 and AP2M were 164 and 171 bp, respectively. The PCR efficiencies of AtHSP90.1 and AP2M were 1.01 and 0.93, respectively. Each sample was measured in triplicate from four independent cDNAs. Amplicon identity was verified by melting curve analysis.
Cloning, Expression, and Purification of HSP90 Proteins
For initial experiments, we expressed and purified the Saccharomyces cerevisiae ScHsp82 protein. The HSP82 coding region was amplified using the primer pair HSP82-1fwd (5’-CGCGGATCCATGGCTAGTGAAACTTTTG-3’) and HSP82-2rev (5’-GCGCGAAGCTTCTAATCTACCTCTTCCATT-3’), which contain BamHI and HindIII sites (underlined), respectively. The resulting 2,156 bp fragment was digested with BamHI and HindIII and ligated into the expression vector pQE-80L (Qiagen) cleaved with the same enzymes yielding plasmid pGW870. The coding region of AtHSP90.1 (At5g52640) was amplified from cDNA with the oligonucleotide primers 5’-CCCCATATGGCGGATGTTCAGATGGC-3’ and 5’-GGACTAGTGTCGACTTCCTCCATCTTGCTC-3’. The resulting PCR product was digested with NdeI and SpeI (restriction sites on primers underlined) and introduced to pQE-80L (Qiagen), cleaved with the same enzymes to obtain the construct pQE80:AtHSP90.1. The cDNA of wheat HSP90 gene TaHSP90-2 (Triticum aestivum; DQ665784.1) was amplified with 5’-CCCCATATGGCGACGGAGACCGAGACC-3’ and 5’-GGACTAGTGTCGACCTCCTCCATCTTGC-3’ (NdeI and SpeI, respectively) and introduced to pQE-80L. All constructs were verified by sequencing.
For production of 6xHis-tagged fusion proteins, the plasmids were introduced into either the standard expression host BL21 (DE3) or the phosphatase-deficient Escherichia coli strain K894 (garB10 fhuA22 phoA4(Am) phoR79::Tn10 ompF627(T2R) serU132(AS) fadL701(T2R) relA1 pitA10 spoT1 rrnB-2 mcrB1 creC510) obtained from the Yale E. coli stock center (#7785).
Protein production was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) following the supplier’s instructions. Protein purification was performed on an Äkta system (GE Healthcare, Uppsala, Sweden). The recombinant proteins were purified by immobilized metal affinity chromatography on Ni2+ charged Chelating Sepharose (GE Healthcare) and eluted with 150 mM of imidazole. Further purification was done by anion exchange chromatography on a 1 ml Resource Q column (GE Healthcare). Proteins were bound to the column in 25 mM Tris/Cl pH 7.5 and eluted with a linear gradient of 0–1 M NaCl in 15 column volumes. The obtained fractions were desalted using a Sephadex™ G-25 fine column, and the purified recombinant proteins were stored in the final elution buffer (25 mM Tris/Cl pH 7.5) at 4°C.
ATPase Activity Assay
The ATPase activities of recombinant proteins AtHSP90.1, TaHSP90-2, and ScHsp82 were assayed following the method of Rowlands et al. (2004) using an ATP concentration of 1 mM. The effect of several potential inhibitors was tested in 96-well plates using toxin concentrations ranging from 1 to 150 µM. All compounds were dissolved in 20% DMSO and further diluted. The assays were performed in a total volume of 25 µl with a final protein concentration of 0.2 µg/µl and 4% DMSO. Controls were incubated with the solvent DMSO without inhibitor. The reactions were incubated at 37°C for 3 h in the dark. After this time, the released inorganic phosphate was quantified with the malachite green colorimetric assay (Rowlands et al., 2004). The reaction was stopped by adding 10 µl of 34% sodium citrate to each well after 20 min incubation at room temperature, and the absorbance was measured at 620 nm.
Production of RAD and RAD-Glucoside
For the production of RAD, baby food jars were filled with 10 g “Langkorn” rice and 10 ml of NaCl solution (100 mg NaCl per jar) and then autoclaved. For inoculation, 1 ml suspension containing about 3 × 106 spores of N. radicicola (strain MA3441 of the Austrian Center of Biological Resources, http://acbr-database.boku.ac.at/) was spread on the rice, and the cultures were incubated at 20°C in the dark for 2 weeks. Then 25 ml of ethanol was added, and after homogenization with an IKA Ultra-Turrax, the extract (containing up to 2.7 g/L RAD) was filtered through Whatman® paper filters. The solvent was partially removed using a rotary evaporator (Buchi R210 Rotavap) at 38°C. To 100 ml of concentrated sample, 25 ml of ethyl acetate (EtOAc, J.T. Baker) was added, and after liquid–liquid partition, the organic phase was concentrated again. The solution was heated to approximately 60°C and slowly cooled down over a period of 3 days in the cold room to crystallize RAD. The residual solvent was removed by vacuum filtration and the crystals washed three times with Milli-Q ultrapure water (Millipore, Molsheim, France). For further purification, preparative HPLC (1100 series, Agilent Technologies, Waldbronn, Germany) was applied. For HPLC separation, a semi-preparative reversed-phase column, Phenomenex Gemini C18, 250 × 10 mm, 5 µm; flow rate: 16 ml/min; eluent A: 90:10 (v:v) Milli-Q H2O:methanol (MeOH) and eluent B: 100% MeOH (LC grade, Merck, Darmstadt, Germany); and injection volume of 500 µl were used. The column was equilibrated with 10% MeOH, which was held for 1 min. A linear gradient was run from 10% to 100% MeOH within 10 min and a hold time of 3 min.
For RAD-glucoside preparation, the genetically modified yeast strain YGZA515 transformed with plasmid pBP918 (Poppenberger et al., 2006), leading to expression of AtUGT73C6, was employed. First, 30 L of log phase cells (OD600 1.1) was produced in synthetic complete medium without leucine (SC-LEU) medium in a bioreactor (Applikon, pH 4, 30°C, 100% oxygen saturation, 10 L/oxygen per hour, and stirring at 600 rpm). The yeast was harvested by centrifugation and the cell paste was transferred into 5 L of fresh SC-LEU medium in a small batch fermenter (pH 5, 30°C, 4.5 g/L glucose, 10 L/oxygen per hour, stirring at 400 rpm). A short heat shock (37°C) was applied prior to RAD addition to induce the expression of stress response genes. A total amount of 18.2 mg RAD was fed to the yeast culture in three equal portions in 1 h intervals. After 46 h, the cells were pelleted by centrifugation, and the supernatant was extracted with an equal amount of EtOAc. The organic phases were collected, transferred into a 1,000 ml rotating flask, and evaporated using a rotary evaporator (Buchi R210 Rotavap) at 40°C. The residue was taken up in 50 ml MeOH and filtrated. The concentrated extract was cleaned of impurities by column chromatography on silica gel (Merck®) using a glass tube with a diameter of 30 mm and a height of 50 cm (silica gel bed 20 cm) using 100% EtOAc to 100% MeOH as mobile phases. Fractions containing RAD-glucoside were concentrated and selected for further purification. Preparative HPLC (1100 series, Agilent Technologies, Waldbronn, Germany) was employed using a semi-preparative column (Phenomenex Gemini C18, 250 × 10 mm, 5 µm). The column temperature was held isothermally at 25°C, and the flow rate was kept constant at 16 ml/min. The injection volume was 500 µl, and the mobile phases were A: MeOH:H2O (20:80, v:v) and B: 100% MeOH LC grade (Merck, Darmstadt, Germany). The chromatography was achieved using binary gradient elution and initiated with MeOH:H2O (20:80) for 2 min, and thereafter, the MeOH content was increased to 100% over 12 min, a hold time of 4 min at the same composition, and then back to the initial conditions over a period of 4 min. RAD-glucoside could be detected by its UV signal at 264 nm. RAD-glucoside–containing fractions were pooled, the solvents were evaporated, and the residue transferred into an 8 ml dark glass vial, evaporated to dryness under N2 and weighed (4.78 mg) using a microbalance (MECAPLEX Switzerland). The purity of RAD-glucoside was tested using an Agilent 1100 HPLC system with a DAD detector [mobile phase aqueous ACN 0%→100%, UV at 200 nm, column: Phenomenex Gemini C18 (150 × 4.6 mm, 5 µm)]. The sum formula of the purified compound was verified by high-resolution mass spectrometry (HR-MS) on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) after negative electrospray ionization.
Results
ZEN Induces the Expression of ATHSP90.1
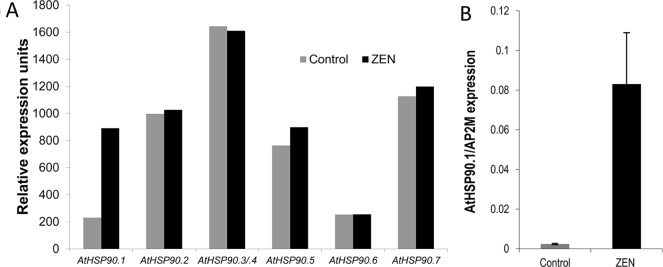
The transcriptional responses of A. thaliana after 2 and 24 h treatments with 50 µM ZEN (corresponding to 15.9 mg/L) were previously studied (Werner, 2005). Besides putative detoxification enzymes (e.g., ABC transporters and UDP-glucosyltransferases), several types of (small) HSPs were found upregulated in this study. In A. thaliana, the HSP90 family consists of seven genes: the encoded proteins are located in the cytosol (AtHSP90.1 to AtHSP90.4), in chloroplasts (AtHSP90.5), in mitochondria (AtHSP90.6), or in the endoplasmic reticulum (AtHSP90.7) (Krishna and Gloor, 2001). AtHSP90.1 is highly stress inducible and of particular importance for disease resistance (Takahashi et al., 2003). It has previously been reported that treatment of A. thaliana seedlings with RAD induced the AtHSP90.1 promoter (Yamada et al., 2007). According to the microarray results, AtHSP90.1 was induced about 3.7-fold after 2 h, while the other members were seemingly unaffected ( Figure 2A ). Using the stable housekeeping gene AT5G46630 as a reference (Wang et al., 2014), quantitative real-time PCR revealed a 35-fold (t-test, p = 0.00372) induction of AtHSP90.1 by ZEN in comparison to the mock control DMSO ( Figure 2B ).
Figure 2.
Transcriptional response of Arabidopsis thaliana HSP90 genes to zearalenone (ZEN). (A) Expression levels of A. thaliana HSP90 genes after 2 h treatment with ZEN (50 µM) or solvent (control; dimethyl sulfoxide, DMSO) according to microarray data from Werner (2005). The genes HSP90.3 and HSP90.4 cannot be distinguished by the probes on the chip (labeled HSP90.3/4). (B) Quantitative real-time PCR of A. thaliana HSP90.1 after 2 h treatment with ZEN (50 µM) or DMSO. The results are expressed as relative expression to the reference gene ADAPTOR PROTEIN-2 MU-ADAPTIN (AP2M; AT5G46630) and represent the means of four biological replicates ± standard deviation.
ZEN and βZEL Inhibit ATPase Activity of HSP90
According to the model proposed by Yamada and Nishimura (2008), inhibition of HSP90 ATPase activates the heat shock transcription factor and consequently induces various HSP genes. This indicates that ZEN could be an HSP90 ATPase inhibitor. To test this hypothesis, we determined the release of phosphate from ATP by HSP90 using the malachite green colorimetric method as described by Rowlands et al. (2004). We first expressed and purified HSP90 from baker’s yeast, ScHsp82p, in E. coli and observed ATPase inhibition for the positive control RAD, and also for ZEN and its phase I metabolites ( Figure 3 ). An IC50 value of about 1.5 µM was calculated for RAD, which is close to the value of 0.9 µM reported by Rowlands et al. (2004). Compared to that, ZEN caused a moderate inhibition of ScHsp82p, with 59 ± 5% (t-test, p = 0.0096) of activity at 100 µM. αZEL did not elicit a detectable response, but ßZEL effectively inhibited ScHsp82p with an IC50 of about 15 µM, resulting in an activity reduction to 8.6 ± 2.4% (p = 0.0016) at 100 µM.
Figure 3.
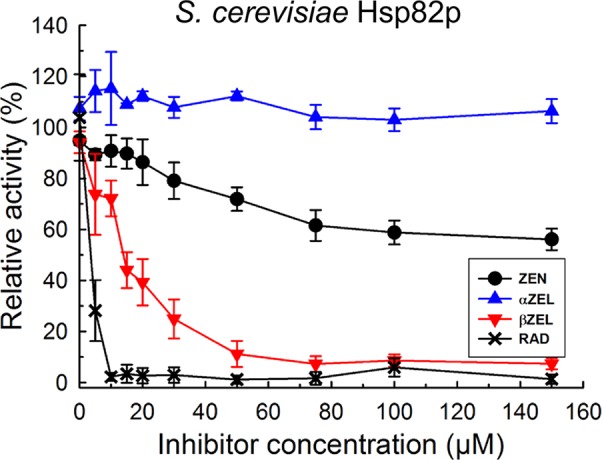
Inhibition of purified 6xHIS-tagged yeast Hsp82p ATPase activity by radicicol (RAD), ZEN, zearalenol-α (αZEL), and zearalenol-β (ßZEL).
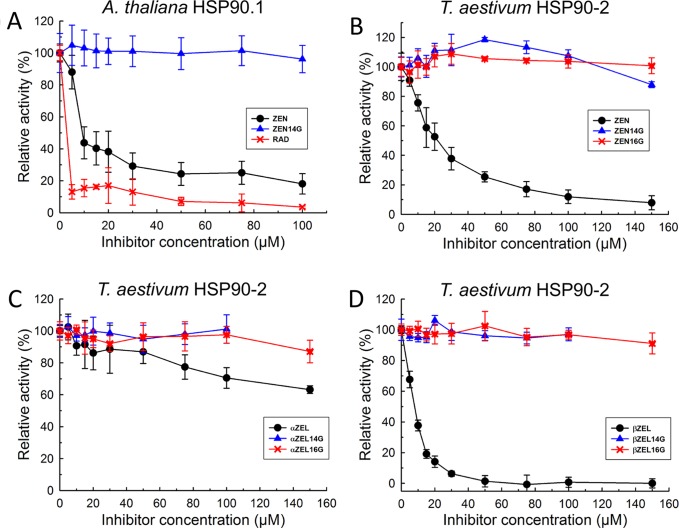
To test whether the yeast model reflects the situation in dicot and monocots plants, AtHSP90.1 and an HSP90 protein from wheat were expressed and purified. The wheat cDNA previously named “TaHSP90-2” corresponds to UniProtKB Q0Q0I7 (Q0Q0I7_WHEAT). This gene belongs to a family of nine highly similar cytosolic HSP90 proteins of wheat, has been named HSP90.3-D1 in a functional study (Wang et al., 2011), and is located on chromosome 5D (= EnsemblPlants Traes_5DL_89CF7F5DE). The results of the ATPase inhibition tests of AtHSP90.1 ( Figure 4A ) and TaHSP90-2 ( Figure 4B ) indicate that ZEN inhibits these more effectively than the yeast protein ( Figure 3 ). At 100 µM ZEN, ATPase activity of AtHSP90.1 was reduced to 18 ± 6% (p = 0.0053) with an IC50 of about 10 µM, and that of TaHSP90-2 to 12 ± 5% (p < 0.0001) with an IC50 of 20 µM. As observed with ScHsp82p, ßZEL had a stronger inhibitory effect than ZEN ( Figure 4D ), causing activity reduction to 0.7 ± 3.3% (p > 0.0001) at 100 µM (IC50 ≈ 10 µM), while αZEL showed low inhibitory activity (71 ± 6% residual ATPase activity, p = 0.0008) at 100 µM ( Figure 4C ).
Figure 4.
Inhibition of Arabidopsis thaliana HSP90.1 (A) and Triticum aestivum HSP90-2 (B–D) ATPase activity by RAD, ZEN, zearalenone-14-glucoside (ZEN14G), zearalenone-16-glucoside (ZEN16G), αZEL, α-zearalenol-14-glucoside (αZEL14G), α-zearalenol-16-glucoside (αZEL16G), ßZEL, ß-zearalenol-14-glucoside (ßZEL14G), and ß-zearalenol-16-glucoside (ßZEL16G).
Glucosylation Abolishes the Inhibitory Activity of RAD, ZEN, and ZELs on HSP90
ZEN is known to be converted to glucosides in planta; therefore, we tested whether glycosylation of ZEN, α/ßZEL and RAD interferes with HSP90 ATPase inhibition. Previously, we had shown that AtUGT73C6, when expressed in a multiple-ABC-transporter–deficient yeast strain, leads to the production of ZEN-14-glucoside [old nomenclature ZEN-4-glucoside, Metzler (2011)] after ZEN treatment (Poppenberger et al., 2006). RAD is toxic for this yeast strain, but the expression of the glucosyltransferase AtUGT73C6 clearly increased RAD resistance ( Figure 5 ). We therefore generated RAD-glucoside for in vitro tests. RAD was purified from N. radicicola and was then used to treat yeast cultures over-expressing AtUGT73C6. In 50 ml overnight cultures, the formation of a compound consistent with the mass of a RAD-glucoside was detected ( Figure 6 ). Since RAD has limited solubility in medium, we produced RAD-glucoside by adding 18 mg RAD to yeast in a small fermenter (see Materials and Methods). The formed RAD-glucoside was purified (4.8 mg) and characterized by liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) and 1H-NMR. The MS/MS fragmentation pattern was consistent with a RAD-glucoside ( Figure 6C ). HR-MS measurements yielded the [M-H]− ion of RAD-glucoside with m/z of 525.1162, confirming the sum formula of C24H27ClO11 (Δm = −1.36 ppm). Unfortunately, like the parent compound RAD, the glucoside appears unstable, particularly when brought to dryness, so it was not possible to obtain a useful NMR spectrum without a prominent water peak. Therefore, we were unable to determine at which position RAD was glucosylated.
Figure 5.
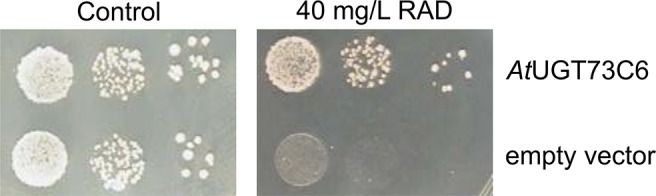
Expression of Arabidopsis thaliana UDP-glucosyltransferase AtUGT73C6 confers radicicol (RAD) resistance to yeast strain YZGA515. Spotting of YZGA515 transformants (three serial 10−1 dilutions) expressing either AtUGT73C6 (upper row) or containing the empty vector. Synthetic complete medium without leucine (SC-LEU) plates without added RAD (left) or with 40 mg/L RAD.
Figure 6.
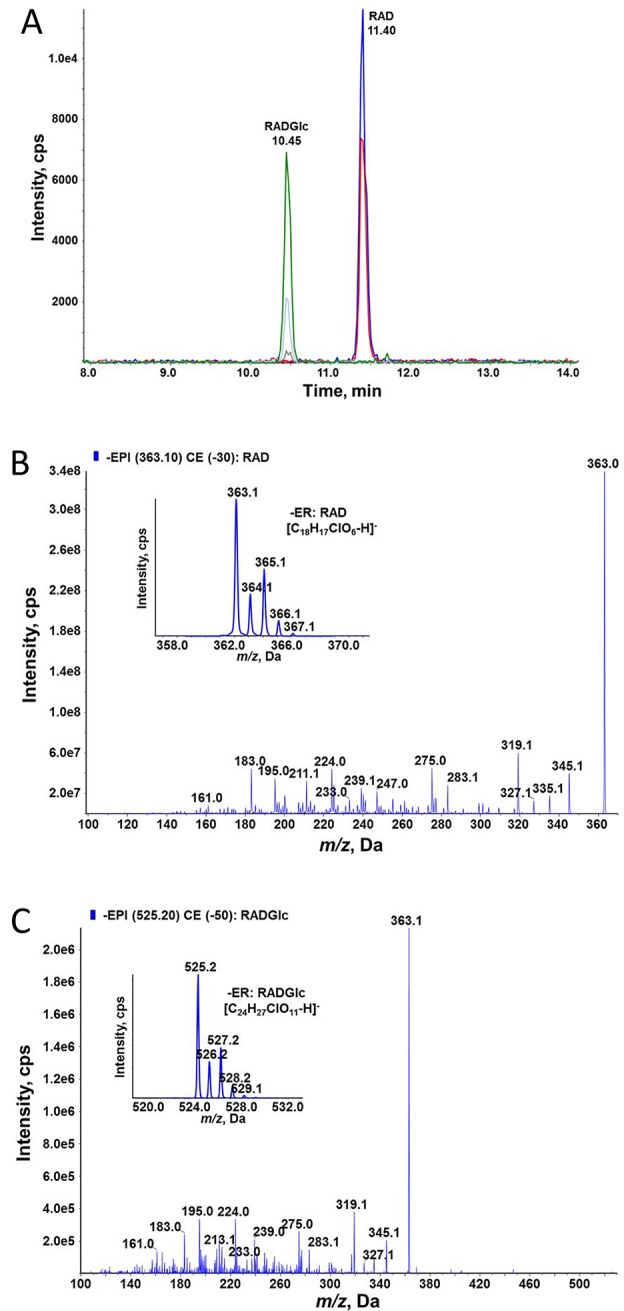
High-performance liquid chromatographic–tandem mass spectrometric (HPLC–MS/MS) determination of RAD and radicicol-glucoside (RADGlc expressing UGT73C6 treated with RAD. (A) HPLC–MS/MS analysis in selective reaction monitoring mode of a culture filtrate of yeast. (B) Enhanced product ion (EPI) MS/MS spectrum of RAD at 30 eV collision energy; the inlay shows the MS spectrum of the deprotonated ions. (C) EPI MS/MS spectrum of RADGlc at 50 eV collision energy; the inlay shows the MS spectrum of the deprotonated ions.
Since AtUGT73C6 produced exclusively ZEN-14-O-glucoside (Poppenberger et al., 2006), we presume RAD was glucosylated at the analogous position. The freshly prepared RAD-glucoside (97% purity) was used to test the activity as an HSP90 inhibitor with recombinant ScHsp82 protein. We observed no inhibitory activity even at the highest tested concentration of 15 µM, demonstrating that glycosylation of RAD abolishes its activity as an HSP90 ATPase inhibitor. Due to the observed instability of the RAD-glucoside, its stability under assay conditions was evaluated by LC–MS/MS quantification before and after the incubation period. This showed no significant (p = 0.21) change in RAD-glucoside concentrations. Likewise, we found all glucosides of ZEN, αZEL, and βZEL stable under identical conditions, with recoveries of > 88%, p > 0.05, in each case.
While ZEN was clearly inhibitory to AtHSP90.1 ( Figure 4A ), ZEN14G was inactive (p = 0.75, 100 µM). Likewise, both ZEN14G and ZEN16G did not cause activity reduction of TaHSP90-2 ( Figure 4B , p > 0.1). The results of inhibition tests with the recombinant wheat TaHSP90-2 are displayed in Figures 4C, D , showing that the 14- and 16-glucosides of both α- and βZEL were inactive as inhibitors and did not cause significant activity reduction. We therefore conclude that glycosylation is an effective detoxification mechanism with respect to the target HSP90.
Discussion
In contrast to the mycotoxin and known virulence factor deoxynivalenol (DON), ZEN is formed only late during the infection process. For wheat, cool and rainy weather delaying harvest is a main factor for ZEN contamination that exceeds maximum tolerated levels (Edwards, 2011). Lack of late season moisture typically leads to low ZEN levels even when Fusarium head blight severity and DON contamination are high (Kharbikar et al., 2015). Also in corn, where ZEN contamination is more frequent and widespread than in small grain cereals, ZEN accumulates much later than DON during infection of corn cobs (Oldenburg and Ellner, 2015). A recent study employing multiple corn inbred lines revealed clear differences between wild-type and ZEN-deficient Fusarium knockout mutants in the employed stalk rot assays with some corn cultivars but not with others (Quesada-Ocampo et al., 2016). The disease symptoms were reduced by about 40%, and also, DON levels were significantly reduced in the stem sections of certain cultivars infected with the ZEN-deficient knockout mutants. In one sweet corn cultivar, the reduction of virulence of the ZEN-deficient mutant was even larger than that observed for a tri5 mutant unable to produce DON. This study suggests that ZEN may have a role in virulence after all, but that corn genotypes may differ in their sensitivity to ZEN.
In order to improve our understanding of Fusarium spp. mycotoxins and their role in plant disease, it is important to identify the mode of action and the molecular targets of these metabolites in planta. The transcriptome response of the model plant A. thaliana gave a first clue, revealing that AtHSP90.1 and several other small HSPs were rapidly and highly induced in response to ZEN treatment (Werner, 2005). Together with the structural similarity to RAD, this led to the hypothesis that ZEN targets HSP90. In this work, we could clearly demonstrate that ZEN and, even more so, its phase I metabolite ßZEL are inhibitors of ATPase of monocot and dicot plants and yeast HSP90 proteins. In A. thaliana (Berthiller et al., 2006), wheat (Rolli et al., 2018), and various yeasts (Böswald et al., 1995), ZEN is rapidly converted into the stronger HSP90 ATPase inhibitory ßZEL and, to a smaller extent, to the less active αZEL, so it matters which form is preferentially generated by the respective host. Since αZEL has much higher estrogenic activity than ZEN and ßZEL in animals, increasing the conversion of ZEN into αZEL would not be a useful Fusarium resistance breeding strategy, if ZEN is indeed a virulence factor. ZEN and ßZEL are much weaker inhibitors than the renowned compound RAD, but the concentrations needed for high-level inhibition in planta are clearly within reach (100 µM correspond to 31.8 mg/L). Some strains of Fusarium even produce gram amounts of ZEN per kilogram substrate in vitro. Naturally infected corn tissue with ZEN contamination levels exceeding 50 mg/kg were reported (di Menna et al., 1997), with local concentrations in infected areas most likely exceeding this by far. The ppm (mg/kg) levels found in Fusarium-infected plant material are in agreement with a possible selective advantage to inhibit a very abundant target with a prominent role in disease resistance.
In general, inhibition of the HSP90 ATPase activity leads to ubiquitin-proteasome–dependent degradation of misfolded client proteins. It was shown that treatment with the HSP90 inhibitor geldanamycin reduced the protein levels of the (epitope-tagged) Pseudomonas resistance gene products RPM1 and RPS5 in A. thaliana (Holt et al., 2005). The hypersensitive response triggered by Pseudomonas syringae DC3000 (avrRpt2) on A. thaliana plants containing the corresponding RPS2 resistance gene was diminished with 10 μM geldanamycin (Takahashi et al., 2003). Many products of classical disease resistance genes are client proteins of HSP90 and its co-chaperones SGT1 and RAR1 (Shirasu, 2009; Kadota et al., 2010; Kadota and Shirasu, 2012). Downregulation of HSP90 gene expression through virus-induced gene silencing or pharmacological inhibition of HSP90 activity led to the breakdown of several gene-for-gene resistance interactions (Shirasu, 2009). It has been shown that virus-induced gene silencing of wheat HSP90 compromises the resistance response to the stripe rust fungus Puccinia striiformis f. sp. tritici. One example requiring HSP90 is the product of the tomato I-2 gene conferring resistance to F. oxysporum (de la Fuente van Bentem et al., 2005). Since HSP90 is a very prominent target in plant–pathogen interaction, it is surprising that loss of ZEN production in Fusarium does not lead to clearly reduced virulence (on the limited number of plants and cultivars tested). The A. thaliana microarray data (Werner, 2005) suggest that several detoxification mechanisms, such as ABC transporters, glutathione-S-transferases, and UDP-glucosyltransferases are induced, which could efficiently counteract the Fusarium small molecule effector ZEN targeting HSP90. Our finding that the ability to inhibit HSP90 is efficiently blocked by glycosylation suggests that ZEN may be a (nearly) defeated virulence factor, neutralized during coevolution. Nevertheless, this effect could be only partial, and genetic differences in substrate specificity and expression levels of certain relevant glucosyltransferases may exist in the breeding material. Yet, the situation is very complex, as diploid plant genomes contain about 180 UGT genes (Caputi et al., 2012). We have not tested RAD metabolism in plants, but the experiment with yeast expressing the A. thaliana UGT73C6 suggests that this compound is also rapidly inactivated into RAD-glucoside in planta. This could also be a reason why geldanamycin instead of RAD was used in previous plant studies.
It has been shown that RAD interacts with human and yeast HSP90 with a highly conserved aspartic acid residue (D79 in Hsp82), mediated by water via hydrogen bonds (Janin, 2010). The interacting parts of RAD are the hydroxyl groups corresponding to the C-14-OH and C-16-OH in ZEN and the carbonyl oxygen in the lactone. It is therefore not surprising that addition of a bulky glucose molecule to either the C-14 or the C-16 hydroxyl group is sufficient to prevent the interaction with the target. These mentioned structural features are the hallmark of metabolites of the class of resorcylic acid lactones, which together with the similar dihydroxy phenyllactic acid lactones form the group of benzenediol lactones, which are extremely widespread metabolites in (plant pathogenic) fungi, particularly in Aigialus, Cochliobolus, Curvularia, Fusarium, Humicola, Lasiodiplodia, Penicillium, and Pochonia species. A recent review lists 190 compounds of this class (Shen et al., 2015). Potentially, several of these metabolites could interact with HSP90 and play a role in plant–pathogen interaction. For instance RAD is also produced by the corn pathogen Colletotrichum graminicola. Yet, also other proteins with a conserved Bergerat-fold ATP binding site could be targeted. One could also speculate that the diversity of this group of fungal metabolites may be driven by the pressure to escape inactivation by glycosylation or analogous phase II detoxification reactions.
While glycosylation is a detoxification mechanism in plants, it has the consequence that animals and humans consuming contaminating grain are also confronted with glucosides and derived substances [e.g., malonylglucosides, di- and tri-glucosides (Rolli et al., 2018)] which are considered masked mycotoxins, as they are not routinely measured but can be hydrolyzed (Gareis et al., 1990; Binder et al., 2017; Dellafiora et al., 2017; Yang et al., 2018b) back to the parental toxin in the intestinal tract of animals and humans. This may increase the actual mycotoxin burden of populations that, based on measured ZEN, already have a high intake (Rai et al., 2018). Besides its estrogenic activity, ZEN also showed pleiotropic toxic effects in various cell lines and experimental animals when applied in high concentrations (Yang et al., 2018a; Zheng et al., 2018; Cao et al., 2019). It is likely that also, HSP90 of humans and animals is inhibited by ZEN, and the resulting effects on multiple client proteins lead to complex pleiotropic toxicological readouts. HSP90 has not been recognized as a ZEN target in transcriptome studies with mammalian cell lines, which may have the trivial reason that the basal level of HSP90 is already so high that the induced level does not reach a typical cutoff of twofold log2. Also in studies where upregulation of HSP90 (and other HSPs) was noticed (e.g., Hassen et al., 2007), this was attributed to increased oxidative stress triggered by ZEN.
Apart from a role as a virulence factor in plant disease development, ZEN may also have an ecological role in preventing competing fungi from colonizing substrates occupied by ZEN producers and preventing mycoparasitism (Utermark and Karlovsky, 2007) or deterring fungivorous soil organisms. It has been reported that mycoparasitic fungi, e.g., Gliocladium roseum (Utermark and Karlovsky, 2007), Sphaerodes mycoparasitica (Kim and Vujanovic, 2017), or Clonostachys rosea (Kosawang et al., 2014), and also other fungi such as Rhizopus species (Brodehl et al., 2014) can counteract ZEN toxicity by formation of ZEN-sulfate, opening the lactone ring, or even by glycosylation.
We could demonstrate here that ZEN targets HSP90 by inhibiting ATPase activity but that this inhibitory activity is effectively antagonized by glycosylation. The taxonomic distribution of ZEN/RAD production in Fusarium spp. and other Hypocreales suggests that these are ancient metabolites (O’Donnell et al., 2013). It is likely they were neutralized to a large extent by plant glycosylation or similar defense responses in the coevolution between pathogens and plants. This might explain why a virulence function of ZEN is difficult to demonstrate, despite its prominent target with an important role in plant defense.
Author Contributions
JATA, HM, and GW constructed plasmids and purified recombinant proteins. JATA and HM performed the ATPase activity assay. WS performed the yeast assay and, together with MS, optimized RAD production. MS purified RAD-glucoside, and EV and FB generated and purified the ZEL-16-O-glucosides. EV, FB, and CH analytically characterized the purified glucosides. M-TH, UW, and DM obtained and analyzed A. thaliana HSP90 expression data. GA and RM initially conceived the idea and obtained first preliminary results. GA, RK, M-TH, and FB obtained funding and supervised the experimental work. JATA and HM prepared figures and, together with GA, wrote the draft manuscript, which was commented on, corrected, and finally approved by all co-authors.
Funding
This work was funded by the Austrian Science Fund (FWF) special research program SFB Fusarium (F3701, F3702, F3706, F3707, F3708, F3711 and F3715).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Katja Sterflinger (BOKU, Department of Biotechnology, Austrian Center of Biological Resources) for providing N. radicicola and Cylindrocarpon strains for screening and optimization of RAD production, and MSc. Romana Stückler for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01160/full#supplementary-material
References
- Aoki T., Vaughan M. M., McCormick S. P., Busman M., Ward T. J., Kelly A., et al. (2015). Fusarium dactylidis sp. nov., a novel nivalenol toxin–producing species sister to F. pseudograminearum isolated from orchard grass (Dactylis glomerata) in Oregon and New Zealand. Mycologia 107, 409–418. 10.3852/14-213 [DOI] [PubMed] [Google Scholar]
- Berthiller F., Hametner C., Krenn P., Schweiger W., Ludwig R., Adam G., et al. (2009. a). Preparation and characterization of the conjugated Fusarium mycotoxins zearalenone-4 O-β-D-glucopyranoside, α-zearalenol-4 O-β-D-glucopyranoside and β-zearalenol-4 O-β-D-glucopyranoside by MS/MS and two-dimensional NMR. Food Addit. Contam. 26, 207–213. 10.1080/02652030802399034 [DOI] [PubMed] [Google Scholar]
- Berthiller F., Schuhmacher R., Adam G., Krska R. (2009. b). Formation, determination and significance of masked and other conjugated mycotoxins. Anal. Bioanal. Chem. 395, 1243–1252. 10.1007/s00216-009-2874-x [DOI] [PubMed] [Google Scholar]
- Berthiller F., Werner U., Sulyok M., Krska R., Hauser M. T., Schuhmacher R. (2006). Liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) determination of phase II metabolites of the mycotoxin zearalenone in the model plant Arabidopsis thaliana . Food Addit. Contam. 23, 1194–1200. 10.1080/02652030600778728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. B., Schwartz-Zimmermann H. E., Varga E., Bichl G., Michlmayr H., Adam G., et al. (2017). Metabolism of zearalenone and its major modified forms in pigs. Toxins (Basel) 9 (2), 56. 10.3390/toxins9020056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böswald C., Engelhardt G., Vogel H., Wallnofer P. R. (1995). Metabolism of the Fusarium mycotoxins zearalenone and deoxynivalenol by yeast strains of technological relevance. Nat. Toxins 3, 138–144. 10.1002/nt.2620030304 [DOI] [PubMed] [Google Scholar]
- Bottalico A., Logrieco A., Visconti A. (1989). Fusarium species and their mycotoxins in infected corn in Italy. Mycopathologia 107, 85–92. 10.1007/BF00707543 [DOI] [PubMed] [Google Scholar]
- Brodehl A., Moller A., Kunte H. J., Koch M., Maul R. (2014). Biotransformation of the mycotoxin zearalenone by fungi of the genera rhizopus and aspergillus. FEMS Microbiol. Lett. 359, 124–130. 10.1111/1574-6968.12586 [DOI] [PubMed] [Google Scholar]
- Caldwell R. W., Tuite J., Stob M., Baldwin R. (1970). Zearalenone production by Fusarium Species. Appl. Microbiol. 20, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Zhi Y., Xu H., Fang H., Jia X. (2019). Zearalenone causes embryotoxicity and induces oxidative stress and apoptosis in differentiated human embryonic stem cells. Toxicol. In Vitro 54, 243–250. 10.1016/j.tiv.2018.09.020 [DOI] [PubMed] [Google Scholar]
- Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042. 10.1111/j.1365-313X.2011.04853.x [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S., Vossen J. H., de Vries K. J., van Wees S., Tameling W. I., Dekker H. L., et al. (2005). Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 43, 284–298. 10.1111/j.1365-313X.2005.02450.x [DOI] [PubMed] [Google Scholar]
- Dellafiora L., Galaverna G., Righi F., Cozzini P., Dall’Asta C. (2017). Assessing the hydrolytic fate of the masked mycotoxin zearalenone-14-glucoside—a warning light for the need to look at the “maskedome”. Food Chem. Toxicol. 99, 9–16. 10.1016/j.fct.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Desjardins A. E. (2006). Fusarium mycotoxins: chemistry, genetics, and biology. St. Paul. MN, USA: The American Phytopathological Society. [Google Scholar]
- di Menna M. E., Lauren D. R., Hardacre A. (1997). Fusaria and Fusarium toxins in New Zealand maize plants. Mycopathologia 139, 165–173. 10.1023/A:1006863908275 [DOI] [PubMed] [Google Scholar]
- Edwards S. (2011). Zearalenone risk in European wheat. World Mycotoxin J. 4, 433–438. 10.3920/WMJ2011.1293 [DOI] [Google Scholar]
- EFSA Panel on Contaminants in the Food Chain (CONTAM) (2011). Scientific opinion on the risks for public health related to the presence of zearalenone in food. EFSA Journal 9 (6), 2197. 10.2903/j.efsa.2011.2197 [DOI] [Google Scholar]
- European Commission (2006. a). Commission recommendation of 17 August 2006 on the presence of deoxynivalenol, zearalenone, ochratoxin A, T-2 and HT-2 and fumonisins in products intended for animal feeding (Text with EEA relevance) (2006/576/EC). Off. J. Eur. Union L229, 7–9. [Google Scholar]
- European Commission. (2006. b). Commission regulation (EC) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs (text with EEA relevance). Off. J. Eur. Union L364, 5–24. [Google Scholar]
- European Commission (2016). Commission recommendation (EU) 2016/1319 of 29 July 2016 amending Recommendation 2006/576/EC as regards deoxynivalenol, zearalenone and ochratoxin A in pet food (text with EEA relevance). Off. J. Eur. Union L508, 58–60. [Google Scholar]
- Gaffoor I., Trail F. (2006). Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl. Environ. Microbiol. 72, 1793–1799. 10.1128/AEM.72.3.1793-1799.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareis M., Bauer J., Thiem J., Plank G., Grabley S., Gedek B. (1990). Cleavage of zearalenone-glycoside, a “masked” mycotoxin, during digestion in swine. Zentralbl. Veterinarmed. B 37, 236–240. 10.1111/j.1439-0450.1990.tb01052.x [DOI] [PubMed] [Google Scholar]
- Grafenhan T., Johnston P. R., Vaughan M. M., McCormick S. P., Proctor R. H., Busman M., et al. (2016). Fusarium praegraminearum sp. nov., a novel nivalenol mycotoxin–producing pathogen from New Zealand can induce head blight on wheat. Mycologia 108, 1229–1239. 10.3852/16-110 [DOI] [PubMed] [Google Scholar]
- Gromadzka K., Waskiewicz A., Chelkowski J., Golinski P. (2008). Zearalenone and its metabolites: occurrence, detection, toxicity and guidelines. World Mycotoxin J. 1, 209–220. 10.3920/WMJ2008.x015 [DOI] [Google Scholar]
- Harris L. J., Balcerzak M., Johnston A., Schneiderman D., Ouellet T. (2016). Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 120, 111–123. 10.1016/j.funbio.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Hassen W., Ayed-Boussema I., Oscoz A. A., Lopez Ade C., Bacha H. (2007). The role of oxidative stress in zearalenone-mediated toxicity in Hep G2 cells: oxidative DNA damage, gluthatione depletion and stress proteins induction. Toxicology 232, 294–302. [DOI] [PubMed] [Google Scholar]
- Holt B. F., 3rd, Belkhadir Y., Dangl J. L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309, 929–932. 10.1126/science.1109977 [DOI] [PubMed] [Google Scholar]
- Janin Y. L. (2010). ATPase inhibitors of heat-shock protein 90, second season. Drug Discov. Today 15, 342–353. 10.1016/j.drudis.2010.03.002 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K. (2012). The HSP90 complex of plants. Biochim. Biophys. Acta 1823, 689–697. 10.1016/j.bbamcr.2011.09.016 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Guerois R. (2010). NLR sensors meet at the SGT1–HSP90 crossroad. Trends Biochem. Sci. 35, 199–207. 10.1016/j.tibs.2009.12.005 [DOI] [PubMed] [Google Scholar]
- Karsai A., Müller S., Platz S., Hauser M. T. (2002). Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32, 790–796. 10.2144/02324st05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbikar L. L., Dickin E. T., Edwards S. G. (2015). Impact of post-anthesis rainfall, fungicide and harvesting time on the concentration of deoxynivalenol and zearalenone in wheat. Food Addit. Contam. Part A: Chem. Anal. Control 32, 2075–2085. 10.1080/19440049.2015.1084652 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Vujanovic V. (2017). Biodegradation and biodetoxification of Fusarium mycotoxins by Sphaerodes mycoparasitica. AMB Express 7, 145. 10.1186/s13568-017-0446-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. T., Lee Y. R., Jin J., Han K. H., Kim H., Kim J. C., et al. (2005). Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 58, 1102–1113. 10.1111/j.1365-2958.2005.04884.x [DOI] [PubMed] [Google Scholar]
- Kosawang C., Karlsson M., Velez H., Rasmussen P. H., Collinge D. B., Jensen B., et al. (2014). Zearalenone detoxification by zearalenone hydrolase is important for the antagonistic ability of Clonostachys rosea against mycotoxigenic Fusarium graminearum . Fungal Biol. 118, 364–373. 10.1016/j.funbio.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Kovalsky Paris M. P., Schweiger W., Hametner C., Stückler R., Muehlbauer G. J., Varga E., et al. (2014). Zearalenone-16-O-glucoside: a new masked mycotoxin. J. Agric. Food Chem. 62, 1181–1189. 10.1021/jf405627d [DOI] [PubMed] [Google Scholar]
- Krenn P., Berthiller F., Schweiger W., Hametner C., Ludwig R., Adam G., et al. (2007). Production of zearalenone-4-glucoside, α-zearalenol-4-glucoside and b-zearalenol-4-glucoside. Mycotoxin Res. 23, 180–184. 10.1007/BF02946045 [DOI] [PubMed] [Google Scholar]
- Krishna P., Gloor G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana . Cell Stress Chaperones 6, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper-Goodman T., Scott P. M., Watanabe H. (1987). Risk assessment of the mycotoxin zearalenone. Regul. Toxicol. Pharmacol. 7, 253–306. 10.1016/0273-2300(87)90037-7 [DOI] [PubMed] [Google Scholar]
- Lysoe E., Klemsdal S. S., Bone K. R., Frandsen R. J., Johansen T., Thrane U., et al. (2006). The PKS4 gene of Fusarium graminearum is essential for zearalenone production. Appl. Environ. Microbiol. 72, 3924–3932. 10.1128/AEM.00963-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysoe E., Seong K. Y., Kistler H. C. (2011). The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact. 24, 995–1000. 10.1094/MPMI-02-11-0038 [DOI] [PubMed] [Google Scholar]
- Metzler M. (2011). Proposal for a uniform designation of zearalenone and its metabolites. Mycotoxin Res. 27, 1–3. 10.1007/s12550-010-0075-2 [DOI] [PubMed] [Google Scholar]
- Michlmayr H., Varga E., Lupi F., Malachová A., Hametner C., Berthiller F., et al. (2017). Synthesis of mono-and di-glucosides of zearalenone and α-/β-zearalenol by recombinant barley glucosyltransferase HvUGT14077 . Toxins 9, 58. 10.3390/toxins9020058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano G. D., Lopez T. A. (1991). Influence of temperature on zearalenone production by regional strains of Fusarium graminearum and Fusarium oxysporum in culture. Int. J. Food Microbiol. 13, 329–333. 10.1016/0168-1605(91)90092-4 [DOI] [PubMed] [Google Scholar]
- Mirrington R. N., Ritchie E., Shoppee C. W., Sternhell S., Taylor W. C. (1965). Some metabolites of Nectria radicicola Gerlach and Nilsson (syn. Cylindrocarpon radicicola Wr.): the structure of radicicol (monorden). Aust. J. Chem. 19, 1265–1284. 10.1071/CH9661265 [DOI] [Google Scholar]
- Molto G. A., Gonzalez H. H. L., Resnik S. L., Gonzalez A. P. (1997). Production of trichothecenes and zearalenone by isolates of Fusarium spp. from Argentinian maize. Food Addit. Contam. 14, 263–268. 10.1080/02652039709374523 [DOI] [PubMed] [Google Scholar]
- Nathanail A. V., Syvahuoko J., Malachova A., Jestoi M., Varga E., Michlmayr H., et al. (2015). Simultaneous determination of major type A and B trichothecenes, zearalenone and certain modified metabolites in Finnish cereal grains with a novel liquid chromatography–tandem mass spectrometric method. Anal. Bioanal. Chem. 407, 4745–4755. 10.1007/s00216-015-8676-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K., Rooney A. P., Proctor R. H., Brown D. W., McCormick S. P., Ward T. J., et al. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 52, 20–31. 10.1016/j.fgb.2012.12.004 [DOI] [PubMed] [Google Scholar]
- Oldenburg E., Ellner F. (2015). Distribution of disease symptoms and mycotoxins in maize ears infected by Fusarium culmorum and Fusarium graminearum . Mycotoxin Res. 31, 117–126. 10.1007/s12550-015-0222-x [DOI] [PubMed] [Google Scholar]
- Picard D. (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59, 1640–1648. 10.1007/PL00012491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Berthiller F., Bachmann H., Lucyshyn D., Peterbauer C., Mitterbauer R., et al. (2006). Heterologous expression of Arabidopsis UDP-glucosyltransferases in Saccharomyces cerevisiae for production of zearalenone-4-O-glucoside. Appl. Environ. Microbiol. 72, 4404–4410. 10.1128/AEM.02544-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-Ocampo L. M., Al-Haddad J., Scruggs A. C., Buell C. R., Trail F. (2016). Susceptibility of maize to stalk rot caused by Fusarium graminearum deoxynivalenol and zearalenone mutants. Phytopathology 106, 920–927. 10.1094/PHYTO-09-15-0199-R [DOI] [PubMed] [Google Scholar]
- Rai A., Dixit S., Singh S. P., Gautam N. K., Das M., Tripathi A. (2018). Presence of zearalenone in cereal grains and its exposure risk assessment in Indian population. J. Food Sci. 83, 3126–3133. 10.1111/1750-3841.14404 [DOI] [PubMed] [Google Scholar]
- Richardson K. E., Hagler W. M., Jr., Campbell C. L., Hamilton P. B. (1985). Production of zearalenone, T-2 toxin, and deoxynivalenol by Fusarium spp. isolated from plant materials grown in North Carolina. Mycopathologia 90, 155–160. 10.1007/BF00436731 [DOI] [PubMed] [Google Scholar]
- Roe S. M., Prodromou C., O’Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1999). Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 42, 260–266. 10.1021/jm980403y [DOI] [PubMed] [Google Scholar]
- Rolli E., Righetti L., Galaverna G., Suman M., Dall’Asta C., Bruni R. (2018). Zearalenone uptake and biotransformation in micropropagated Triticum durum desf. plants: a xenobolomic approach. J. Agric. Food Chem. 66, 1523–1532. 10.1021/acs.jafc.7b04717 [DOI] [PubMed] [Google Scholar]
- Rowlands M. G., Newbatt Y. M., Prodromou C., Pearl L. H., Workman P., Aherne W. (2004). High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal. Biochem. 327, 176–183. 10.1016/j.ab.2003.10.038 [DOI] [PubMed] [Google Scholar]
- Sarver B. A., Ward T. J., Gale L. R., Broz K., Kistler H. C., Aoki T., et al. (2011). Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 48, 1096–1107. 10.1016/j.fgb.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Schulte T. W., Akinaga S., Soga S., Sullivan W., Stensgard B., Toft D., et al. (1998). Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgobba M., Rastelli G. (2009). Structure-based and in silico design of Hsp90 inhibitors. Chem. Med. Chem. 4, 1399–1409. 10.1002/cmdc.200900256 [DOI] [PubMed] [Google Scholar]
- Shen W., Mao H., Huang Q., Dong J. (2015). Benzenediol lactones: a class of fungal metabolites with diverse structural features and biological activities. Eur. J. Med. Chem. 97, 747–777. 10.1016/j.ejmech.2014.11.067 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90–SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. 10.1146/annurev.arplant.59.032607.092906 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 100, 11777–11782. 10.1073/pnas.2033934100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermark J., Karlovsky P. (2007). Role of zearalenone lactonase in protection of Gliocladium roseum from fungitoxic effects of the mycotoxin zearalenone. Appl. Environ. Microbiol. 73, 637–642. 10.1128/AEM.01440-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee T., Zhang H., van Diepeningen A., Waalwijk C. (2015). Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Addit. Contam. Part A: Chem. Anal. Control 32, 453–460. 10.1080/19440049.2014.984244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. F., Wei X., Fan R., Zhou H., Wang X., Yu C., et al. (2011). Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 191, 418–431. 10.1111/j.1469-8137.2011.03715.x [DOI] [PubMed] [Google Scholar]
- Wang H., Wang J., Jiang J., Chen S., Guan Z., Liao Y., et al. (2014). Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis . Scientific Reports 4, 6781. 10.1038/srep06781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner U. (2005). Characterisation of the effect of the Fusarium mycotoxin zearalenone in Arabidopsis thaliana [dissertation]. Vienna (Austria): University of Natural Resources and Life Sciences. [Google Scholar]
- Yamada K., Fukao Y., Hayashi M., Fukazawa M., Suzuki I., Nishimura M. (2007). Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana . J. Biol. Chem. 282, 37794–37804. 10.1074/jbc.M707168200 [DOI] [PubMed] [Google Scholar]
- Yamada K., Nishimura M. (2008). Cytosolic heat shock protein 90 regulates heat shock transcription factor in Arabidopsis thaliana . Plant Signal Behav. 3, 660–662. 10.4161/psb.3.9.5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Jiang X., Sun J., Li X., Li X., Jiao R., et al. (2018. a). Toxic effects of zearalenone on gametogenesis and embryonic development: a molecular point of review. Food Chem. Toxicol. 119, 24–30. 10.1016/j.fct.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang H., Zhang J., Li Y., Jin Y., Zhang S., et al. (2018. b). Deglucosylation of zearalenone-14-glucoside in animals and human liver leads to underestimation of exposure to zearalenone in humans. Arch. Toxicol. 92, 2779–2791. 10.1007/s00204-018-2267-z [DOI] [PubMed] [Google Scholar]
- Zheng W., Wang B., Li X., Wang T., Zou H., Gu J., et al. (2018). Zearalenone promotes cell proliferation or causes cell death? Toxins (Basel) 10 (5), 184. 10.3390/toxins10050184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinedine A., Soriano J. M., Molto J. C., Manes J. (2007). Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: an oestrogenic mycotoxin. Food Chem. Toxicol. 45, 1–18. 10.1016/j.fct.2006.07.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.